8WD5
 
 | |
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | 分子名称: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | 著者 | Dong, X, Gong, Z, Hu, Y.F. | | 登録日 | 2023-07-14 | | 公開日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
7BTK
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | 分子名称: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | 登録日 | 2020-04-01 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | 分子名称: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | 登録日 | 2020-03-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5J1A
 
 | | Antigen presenting molecule | | 分子名称: | 3-[(8Z,11Z)-pentadeca-8,11-dien-1-yl]benzene-1,2-diol, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | 著者 | Yongqing, T, Rossjohn, J, Le Nours, J, Marquez, E, Winau, F. | | 登録日 | 2016-03-29 | | 公開日 | 2016-08-03 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | CD1a on Langerhans cells controls inflammatory skin disease.
Nat. Immunol., 17, 2016
|
|
3VKD
 
 | | Crystal structure of MoeO5 soaked with 3-phosphoglycerate | | 分子名称: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | 著者 | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | 登録日 | 2011-11-12 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VK5
 
 | | Crystal structure of MoeO5 in complex with its product FPG | | 分子名称: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | 著者 | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | 登録日 | 2011-11-08 | | 公開日 | 2012-05-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKC
 
 | | Crystal structure of MoeO5 soaked with pyrophosphate | | 分子名称: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | 著者 | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | 登録日 | 2011-11-12 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKB
 
 | | Crystal structure of MoeO5 soaked with FsPP overnight | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, MoeO5, ... | | 著者 | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | 登録日 | 2011-11-11 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKA
 
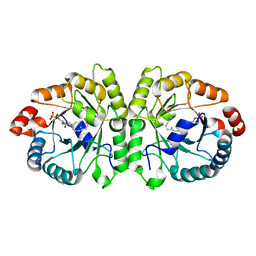 | | Crystal structure of MoeO5 soaked for 3 hours in FsPP | | 分子名称: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | 著者 | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | 登録日 | 2011-11-11 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5YYL
 
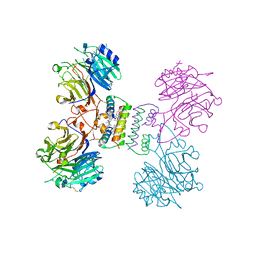 | | Structure of Major Royal Jelly Protein 1 Oligomer | | 分子名称: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Tian, W, Chen, Z. | | 登録日 | 2017-12-10 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
8DKC
 
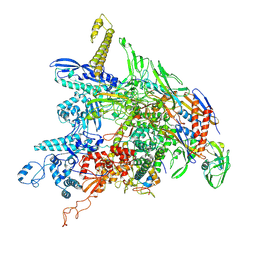 | | P. gingivalis RNA Polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, B, Bu, F. | | 登録日 | 2022-07-05 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
8GZU
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory Megacomplex MC (IV2+I+III2+II)2 | | 分子名称: | 2 iron, 2 sulfur cluster-binding protein, 2-oxoglutarate/malate carrier protein, ... | | 著者 | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | 登録日 | 2022-09-27 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
6JJS
 
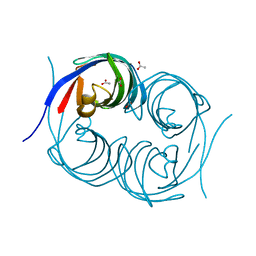 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | 分子名称: | ACETATE ION, PhnH | | 著者 | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-02-26 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6JJT
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | 著者 | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-02-26 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6JKP
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense in complex with NAD+ | | 分子名称: | Methanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.008 Å) | | 主引用文献 | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6JKO
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense | | 分子名称: | Methanol dehydrogenase, ZINC ION | | 著者 | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
2DS2
 
 | | Crystal structure of mabinlin II | | 分子名称: | ACETIC ACID, Sweet protein mabinlin-2 chain A, Sweet protein mabinlin-2 chain B | | 著者 | Li, D.F, Zhu, D.Y, Wang, D.C. | | 登録日 | 2006-06-19 | | 公開日 | 2007-06-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.
J.Struct.Biol., 162, 2008
|
|
8GTY
 
 | |
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | 著者 | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
2G0C
 
 | |
3QW9
 
 | | Crystal structure of betaglycan ZP-C domain | | 分子名称: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Lin, S.J, Jardetzky, T.S. | | 登録日 | 2011-02-27 | | 公開日 | 2011-04-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2HJV
 
 | |
5ZVV
 
 | | Structure of SeMet-phAimR | | 分子名称: | AimR transcriptional regulator, GLYCEROL | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-13 | | 公開日 | 2018-09-05 | | 最終更新日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
