3BAG
 
 | |
3BAQ
 
 | |
3BAU
 
 | |
3BAH
 
 | |
3BAO
 
 | |
3BAV
 
 | |
3BB2
 
 | |
3VY9
 
 | | Crystal structure of PorB from Neisseria meningitidis in complex with cesium ion, space group H32 | | 分子名称: | CESIUM ION, Outer membrane protein | | 著者 | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | 登録日 | 2012-09-21 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3VY8
 
 | | Crystal Structure of PorB from Neisseria meningitidis in complex with Cesium ion, space group P63 | | 分子名称: | CESIUM ION, Outer membrane protein | | 著者 | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | 登録日 | 2012-09-21 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3WI5
 
 | | Crystal structure of the Loop 7 mutant PorB from Neisseria meningitidis serogroup B | | 分子名称: | CITRATE ANION, Major outer membrane protein P.IB | | 著者 | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | 登録日 | 2013-09-05 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
2DVY
 
 | | Crystal structure of restriction endonucleases PabI | | 分子名称: | Restriction endonuclease PabI | | 著者 | Miyazono, K, Watanabe, M, Kamo, M, Sawasaki, T, Nagata, K, Endo, Y, Tanokura, M, Kobayashi, I. | | 登録日 | 2006-08-01 | | 公開日 | 2007-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Novel protein fold discovered in the PabI family of restriction enzymes
Nucleic Acids Res., 35, 2007
|
|
2HWA
 
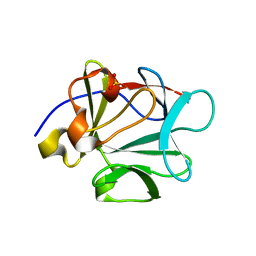 | | Crystal structure of Lys12Thr/Cys117Val mutant of human acidic fibroblast growth factor at 1.65 angstrom resolution. | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2006-08-01 | | 公開日 | 2007-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HW9
 
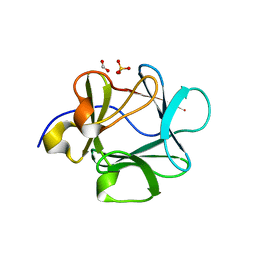 | | Crystal structure of Lys12Cys/Cys117Val mutant of human acidic fibroblast Growth factor at 1.60 angstrom resolution. | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2006-08-01 | | 公開日 | 2007-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HWM
 
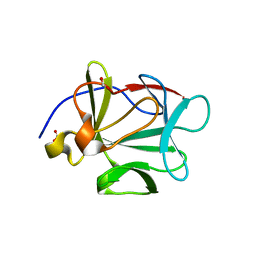 | | Crystal structure of Lys12Val/Cys117Val mutant of human acidic fibroblast growth factor at 1.60 angstrom resolution | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1 | | 著者 | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2006-08-01 | | 公開日 | 2007-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
1UEV
 
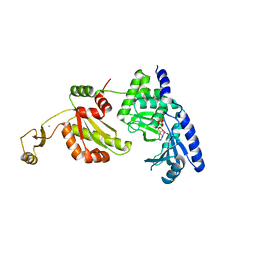 | |
1UET
 
 | |
1UEU
 
 | |
1DYC
 
 | |
1DYD
 
 | |
1DYF
 
 | |
1DYB
 
 | |
1DYE
 
 | |
1DYA
 
 | |
1DYG
 
 | |
2Z6F
 
 | |
