3E1F
 
 | |
5IFM
 
 | | Human NONO (p54nrb) Homodimer | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | 著者 | Knott, G.J, Bond, C.S. | | 登録日 | 2016-02-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3DYP
 
 | | E. coli (lacZ) beta-galactosidase (H418N) | | 分子名称: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Huber, R.E, Matthews, B.W. | | 登録日 | 2008-07-28 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
5G5U
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa | | 分子名称: | BETA-HEXOSAMINIDASE | | 著者 | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | 登録日 | 2016-06-05 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5I0F
 
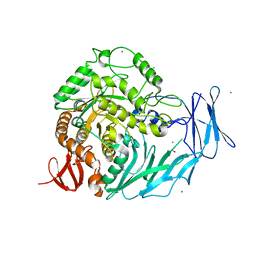 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with covalent intermediate | | 分子名称: | CALCIUM ION, Glycoside hydrolase family 31, TRIETHYLENE GLYCOL, ... | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5J1M
 
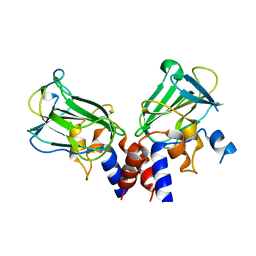 | | Crystal structure of Csd1-Csd2 dimer II | | 分子名称: | ToxR-activated gene (TagE), ZINC ION | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
3DYO
 
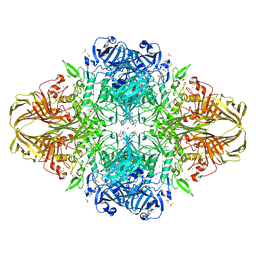 | | E. coli (lacZ) beta-galactosidase (H418N) in complex with IPTG | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Juers, D.H, Huber, R.E, Matthews, B.W. | | 登録日 | 2008-07-28 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
5J1K
 
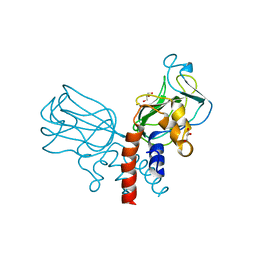 | | Crystal structure of Csd2-Csd2 dimer | | 分子名称: | GLYCEROL, ToxR-activated gene (TagE) | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1L
 
 | | Crystal structure of Csd1-Csd2 dimer I | | 分子名称: | ToxR-activated gene (TagE), ZINC ION | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
1E9Z
 
 | |
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | 分子名称: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | 著者 | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | 登録日 | 2000-11-01 | | 公開日 | 2001-11-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
4IWT
 
 | |
4IVV
 
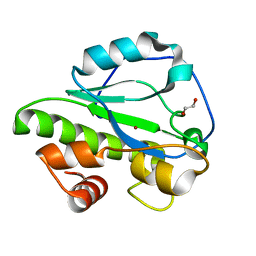 | |
4OZS
 
 | | RNA binding protein | | 分子名称: | Alpha solenoid protein | | 著者 | Gully, B.S, Bond, C.S. | | 登録日 | 2014-02-19 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | The design and structural characterization of a synthetic pentatricopeptide repeat protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CTV
 
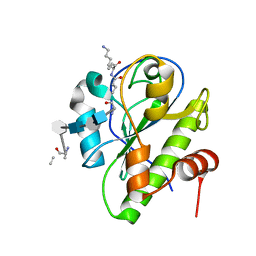 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | 著者 | Achour, A, Sandalova, T, Mellroth, P. | | 登録日 | 2015-07-24 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
5CA5
 
 | |
3U6I
 
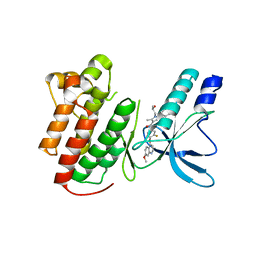 | |
3U6J
 
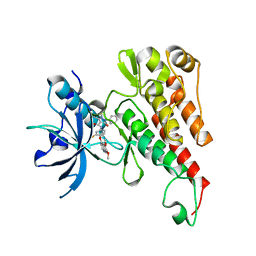 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyrazolone inhibitor | | 分子名称: | N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide, Vascular endothelial growth factor receptor 2 | | 著者 | Whittington, D.A, Long, A, Rose, P, Gu, Y, Zhao, H. | | 登録日 | 2011-10-12 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3U6H
 
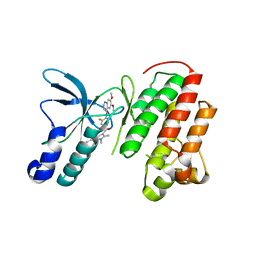 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | 分子名称: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | 著者 | Bellon, S.F, Whittington, D.A, Long, A.L. | | 登録日 | 2011-10-12 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
2BWS
 
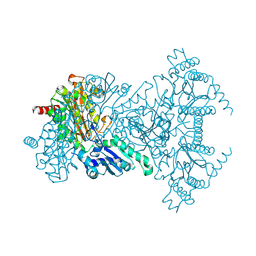 | | His243Ala Escherichia coli Aminopeptidase P | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, XAA-PRO AMINOPEPTIDASE P | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWV
 
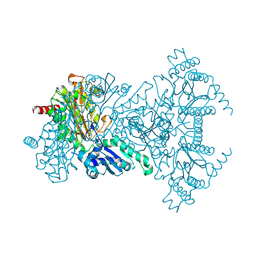 | | His361Ala Escherichia coli Aminopeptidase P | | 分子名称: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWX
 
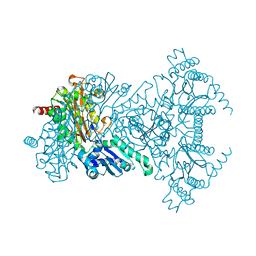 | | His354Ala Escherichia coli Aminopeptidase P | | 分子名称: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWW
 
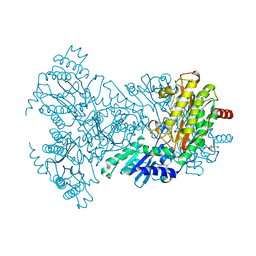 | | His350Ala Escherichia coli Aminopeptidase P | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, AMINOPEPTIDASE P, CITRATE ANION, ... | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWY
 
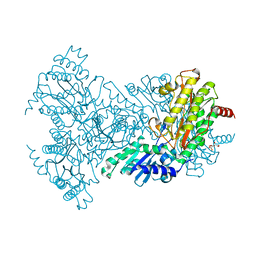 | | Glu383Ala Escherichia coli Aminopeptidase P | | 分子名称: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWU
 
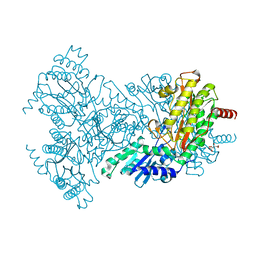 | | Asp271Ala Escherichia coli Aminopeptidase P | | 分子名称: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | 著者 | Graham, S.C, Guss, J.M. | | 登録日 | 2005-07-19 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
