4JY6
 
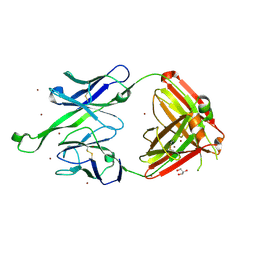 | | Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, PGT123 heavy chain, PGT123 light chain, ... | | 著者 | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans.
Plos Pathog., 9, 2013
|
|
4JM2
 
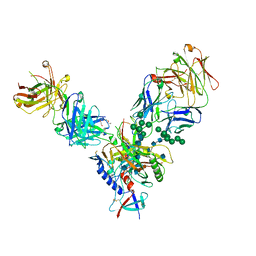 | |
4JM4
 
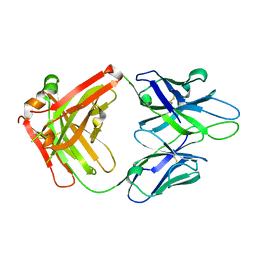 | | Crystal Structure of PGT 135 Fab | | 分子名称: | PGT 135 Heavy Chain, PGT 135 Light Chain | | 著者 | Kong, L, Wilson, I.A. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-29 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4HB9
 
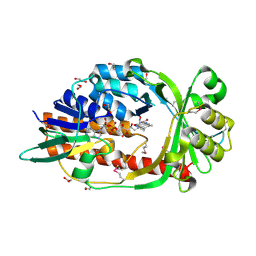 | |
4JY5
 
 | | Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, PGT122 heavy chain, PGT122 light chain | | 著者 | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
7BYO
 
 | |
7BYP
 
 | |
7D04
 
 | |
7D05
 
 | |
7D02
 
 | |
7D01
 
 | |
7EXP
 
 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | 分子名称: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | 著者 | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | 登録日 | 2021-05-28 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
4NCI
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation | | 分子名称: | DNA double-strand break repair Rad50 ATPase | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCJ
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation with ADP Beryllium Flouride | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA double-strand break repair Rad50 ATPase, ... | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCK
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | 分子名称: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
3NR1
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | 分子名称: | HD domain-containing protein 3, MANGANESE (II) ION | | 著者 | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | 登録日 | 2010-06-30 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
4NCH
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation | | 分子名称: | DNA double-strand break repair Rad50 ATPase, SULFATE ION | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4RQQ
 
 | |
3JQ7
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6-phenylpteridine-2,4,7-triamine (DX2) | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-phenylpteridine-2,4,7-triamine, ACETATE ION, ... | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQF
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 1,3,5-triazine-2,4,6-triamine (AX2) | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1,3,5-triazine-2,4,6-triamine, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQ6
 
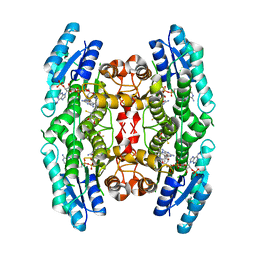 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6,7-bis(1-methylethyl)pteridine-2,4-diamine (DX1) | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6,7-bis(1-methylethyl)pteridine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQE
 
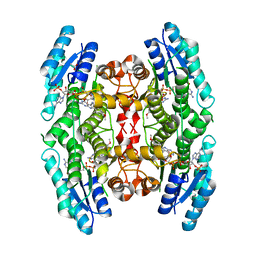 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-(4-methoxyphenyl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (DX8) | | 分子名称: | 2-amino-6-(4-methoxyphenyl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
4P9M
 
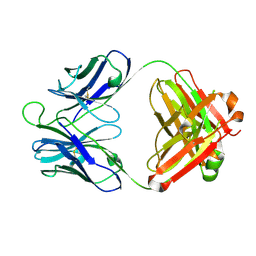 | | Crystal structure of 8ANC195 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, 8ANC195 light chain | | 著者 | Scharf, L, Bjorkman, P.J. | | 登録日 | 2014-04-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike.
Cell Rep, 7, 2014
|
|
3JQ8
 
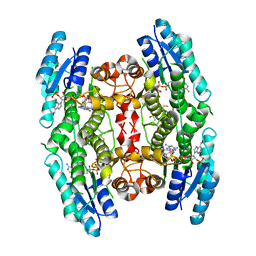 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6,7,7-trimethyl-7,8-dihydropteridine-2,4-diamine (DX3) | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6,7,7-trimethyl-7,8-dihydropteridine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQ9
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (AX1) | | 分子名称: | 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | 著者 | Tulloch, L.B, Hunter, W.N. | | 登録日 | 2009-09-06 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
