6JDU
 
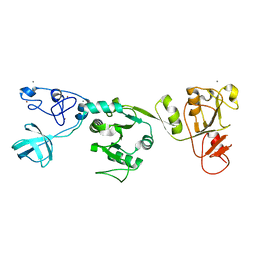 | | Crystal structure of PRRSV nsp10 (helicase) | | 分子名称: | CALCIUM ION, PP1b, ZINC ION | | 著者 | Tang, C, Chen, Z. | | 登録日 | 2019-02-02 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
5YIM
 
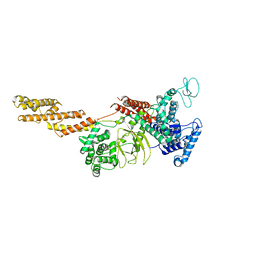 | |
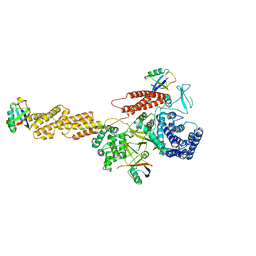
5YIK
 
 | |
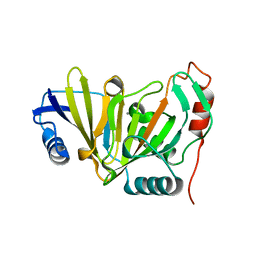
5YIJ
 
 | | Structure of a Legionella effector with substrates | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | 著者 | Feng, Y, Mu, Y, Wang, H. | | 登録日 | 2017-10-05 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
8HB2
 
 | |
8HBB
 
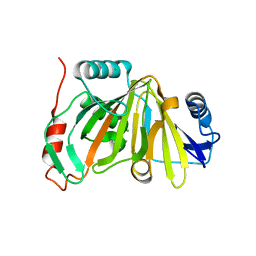 | |
8HAZ
 
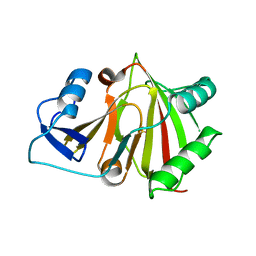 | |
7VXP
 
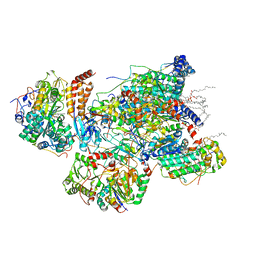 | | Matrix arm of active state CI from Q10 dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VXU
 
 | | Matrix arm of deactive state CI from Q10 dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VY1
 
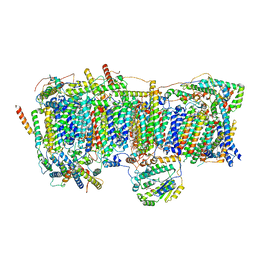 | | Membrane arm of deactive state CI from Q10 dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VXS
 
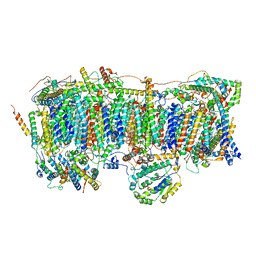 | | Membrane arm of active state CI from Q10 dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W17
 
 | |
7W14
 
 | |
7VYL
 
 | |
7VYK
 
 | |
7VYM
 
 | |
7WG8
 
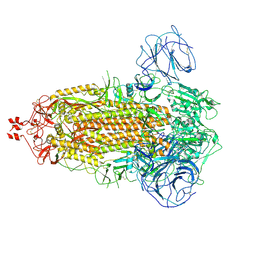 | | Delta Spike Trimer(3 RBD Down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7VYF
 
 | | Matrix arm of active state CI from Rotenone dataset | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4BXF
 
 | |
7VYH
 
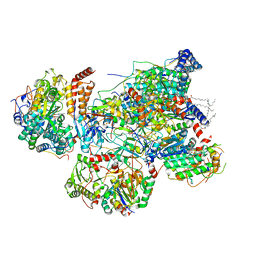 | | Matrix arm of deactive state CI from Rotenone dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VYI
 
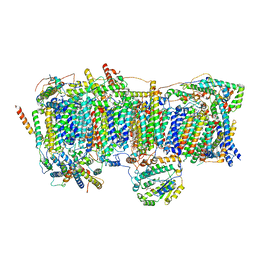 | | Membrane arm of deactive state CI from Rotenone dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6M3M
 
 | |
7WE6
 
 | | Structure of Csy-AcrIF24-dsDNA | | 分子名称: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L, Feng, Y. | | 登録日 | 2021-12-22 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
