5JCL
 
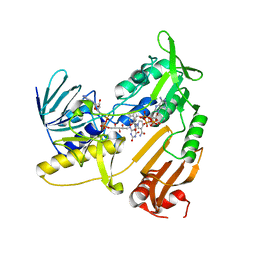 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCK
 
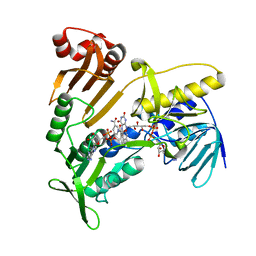 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6
|
|
5JCN
 
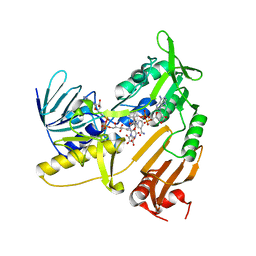 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | ASCORBIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCI
 
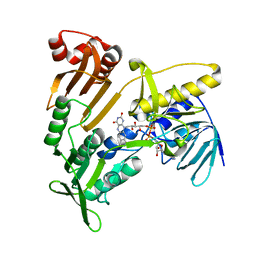 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCM
 
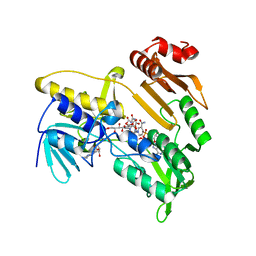 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, ISOASCORBIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
4ES0
 
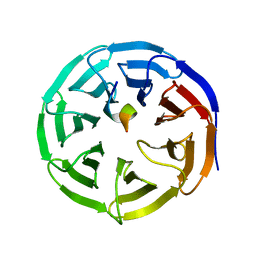 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-21 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.817 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERQ
 
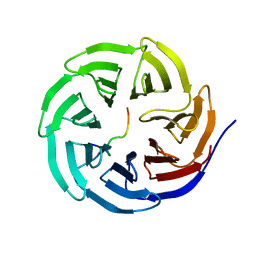 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-20 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ESG
 
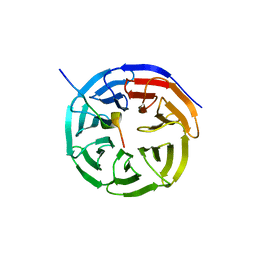 | | X-ray structure of WDR5-MLL1 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-23 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-21 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4EWR
 
 | | X-ray structure of WDR5-SETd1a Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-27 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERZ
 
 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-21 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
3NQW
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | 分子名称: | CG11900, MANGANESE (II) ION, SULFATE ION | | 著者 | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | 登録日 | 2010-06-30 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
6KZB
 
 | | Transglutaminase2 complexed with calcium | | 分子名称: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | 著者 | Park, H.H, Kim, C.M. | | 登録日 | 2019-09-23 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2.
Int J Mol Sci, 21, 2020
|
|
1MNL
 
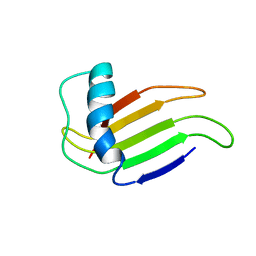 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | 分子名称: | MONELLIN | | 著者 | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | 登録日 | 1998-08-06 | | 公開日 | 1999-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
7YSI
 
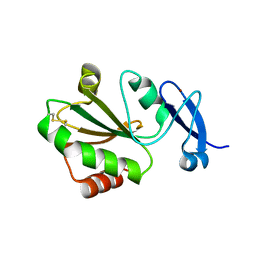 | | Crystal structure of thioredoxin 2 | | 分子名称: | Thiol disulfide reductase thioredoxin, ZINC ION | | 著者 | Chang, Y.J, Park, H.H. | | 登録日 | 2022-08-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.202 Å) | | 主引用文献 | Comparison of the structure and activity of thioredoxin 2 and thioredoxin 1 from Acinetobacter baumannii.
Iucrj, 10, 2023
|
|
4TOY
 
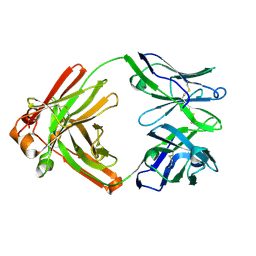 | |
4UC4
 
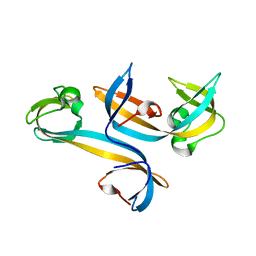 | |
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | 分子名称: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | 著者 | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | 登録日 | 2015-07-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
3MVE
 
 | | Crystal structure of a novel pyruvate decarboxylase | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | 著者 | Cha, S.S, Jeong, C.S, An, Y.J. | | 登録日 | 2010-05-04 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
4JY4
 
 | | Crystal structure of human Fab PGT121, a broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PGT121 heavy chain, ... | | 著者 | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
4JM2
 
 | |
4JM4
 
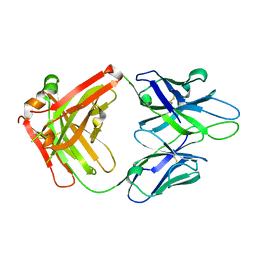 | | Crystal Structure of PGT 135 Fab | | 分子名称: | PGT 135 Heavy Chain, PGT 135 Light Chain | | 著者 | Kong, L, Wilson, I.A. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-29 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JY6
 
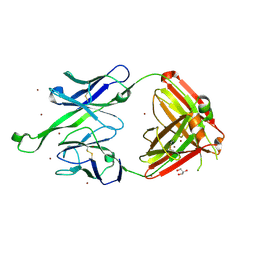 | | Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, PGT123 heavy chain, PGT123 light chain, ... | | 著者 | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans.
Plos Pathog., 9, 2013
|
|
4JY5
 
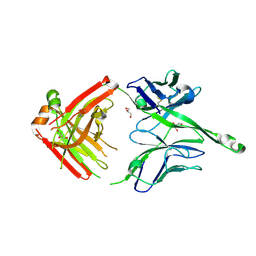 | | Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, PGT122 heavy chain, PGT122 light chain | | 著者 | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
3NR1
 
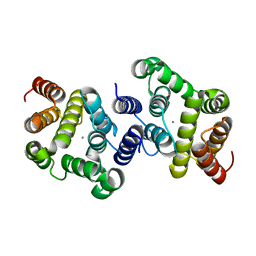 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | 分子名称: | HD domain-containing protein 3, MANGANESE (II) ION | | 著者 | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | 登録日 | 2010-06-30 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
