8QUZ
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on analytical absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUU
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on spherical harmonics absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
4KF7
 
 | |
4KF8
 
 | |
4ODB
 
 | |
6ZWO
 
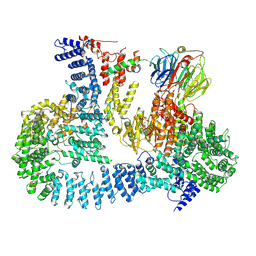 | | cryo-EM structure of human mTOR complex 2, focused on one half | | 分子名称: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | 登録日 | 2020-07-28 | | 公開日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6ZWM
 
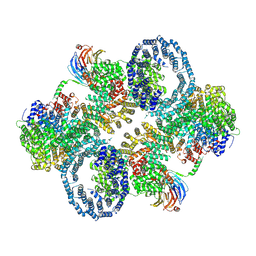 | | cryo-EM structure of human mTOR complex 2, overall refinement | | 分子名称: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | 登録日 | 2020-07-28 | | 公開日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6MPD
 
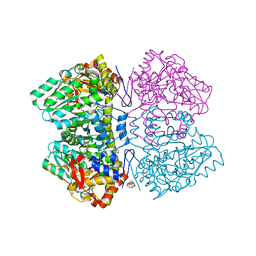 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from 3-F-L-tyrosine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-FLUOROTYROSINE, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-10-05 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MME
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-09-30 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MO3
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-serine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-10-04 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MLS
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-tyrosine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-09-27 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MQQ
 
 | | Citrobacter freundii F448A mutant tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-10-10 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | 分子名称: | GLYCEROL, PCZA361.16 | | 著者 | Merkel, A.B, Naismith, J.H. | | 登録日 | 2003-04-17 | | 公開日 | 2004-04-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3DL1
 
 | |
5ZO0
 
 | |
5ZKZ
 
 | |
5ZIW
 
 | |
2XE2
 
 | |
2XE3
 
 | | OmpC28 | | 分子名称: | OUTER MEMBRANE PORIN C, octyl beta-D-glucopyranoside | | 著者 | Lou, H, Naismith, J.H. | | 登録日 | 2010-05-10 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Altered Antibiotic Transport in Ompc Mutants Isolated from a Series of Clinical Strains of Multi-Drug Resistant E. Coli.
Plos One, 6, 2011
|
|
2XE5
 
 | |
2XG6
 
 | |
2XE1
 
 | |
6K9R
 
 | |
6K9O
 
 | | Crystal Structure Analysis of Protein | | 分子名称: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | 著者 | Li, C, Wan, Q. | | 登録日 | 2019-06-17 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
8AOU
 
 | |
