7QZ5
 
 | |
4WIM
 
 | |
3N4G
 
 | | Crystal structure of native Cg10062 | | 分子名称: | Putative tautomerase | | 著者 | Guo, Y, Robertson, B.A, Hackert, M.L, Whitman, C.P. | | 登録日 | 2010-05-21 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal Structures of the Native and Inactivated Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase from Corynebacterium glutamicum: Implications for the Evolution of cis-3-Chloroacrylic Acid Dehalogenase Activity in the Tautomerase Superfamily
To be Published
|
|
3N4D
 
 | |
3N4H
 
 | |
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | 分子名称: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | 著者 | Sang, Y.S, Cai, M, Clore, G.M. | | 登録日 | 2009-10-29 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WY2
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | 分子名称: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | 著者 | Sang, Y.S, Cai, M, Clore, G.M. | | 登録日 | 2009-11-11 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
1LPF
 
 | |
2B9V
 
 | |
1GUV
 
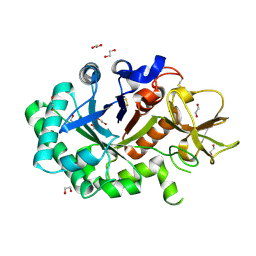 | | Structure of human chitotriosidase | | 分子名称: | 1,2-ETHANEDIOL, CHITOTRIOSIDASE | | 著者 | Von Moeller, H, Houston, D, Boot, R.G, Aerts, J.M.F.G, Van Aalten, D.M.F. | | 登録日 | 2002-01-31 | | 公開日 | 2003-03-30 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of Human Chitotriosidase - Implications for Specific Inhibitor Design and Function of Mammalian Chitinase-Like Lectins
J.Biol.Chem., 277, 2002
|
|
1RYY
 
 | |
2FLT
 
 | |
2FLZ
 
 | |
2GUY
 
 | |
1CNV
 
 | |
3LAD
 
 | |
1B48
 
 | |
1K9T
 
 | | Chitinase a complexed with tetra-N-acetylchitotriose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Prag, G, Tucker, P.A, Oppenheim, A.B. | | 登録日 | 2001-10-30 | | 公開日 | 2002-11-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
1PP2
 
 | |
