3LH3
 
 | | DFP modified DegS delta PDZ | | 分子名称: | Protease degS | | 著者 | Sohn, J, Grant, R.A, Sauer, R.T. | | 登録日 | 2010-01-21 | | 公開日 | 2010-08-25 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
4Q2Z
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | 分子名称: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | 著者 | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | 登録日 | 2014-04-10 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | 分子名称: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | 著者 | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | 登録日 | 2014-07-30 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.676 Å) | | 主引用文献 | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
3MMP
 
 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | 分子名称: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | 著者 | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | 登録日 | 2010-04-20 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O1F
 
 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | 分子名称: | ATP-dependent Clp protease adapter protein clpS | | 著者 | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | 登録日 | 2010-07-21 | | 公開日 | 2011-07-27 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
3NPQ
 
 | |
3O2B
 
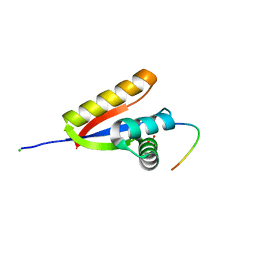 | | E. coli ClpS in complex with a Phe N-end rule peptide | | 分子名称: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | 著者 | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | 登録日 | 2010-07-22 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
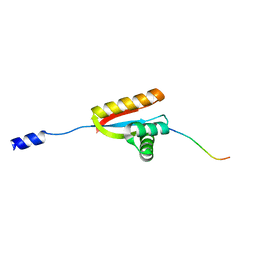 | | E. coli ClpS in complex with a Leu N-end rule peptide | | 分子名称: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | 著者 | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | 登録日 | 2010-07-22 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
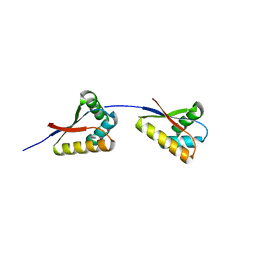 | | Structure of E. coli ClpS ring complex | | 分子名称: | ATP-dependent Clp protease adaptor protein ClpS | | 著者 | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | 登録日 | 2010-07-22 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3NPN
 
 | |
3OF8
 
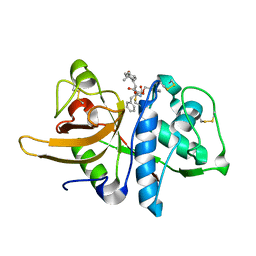 | |
3OF9
 
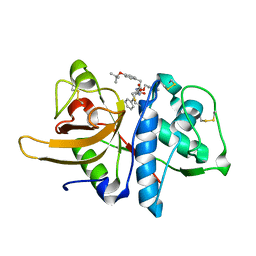 | |
3OTP
 
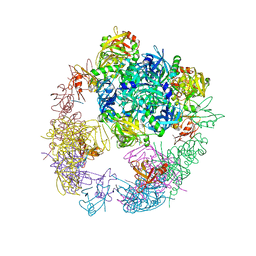 | |
3OU0
 
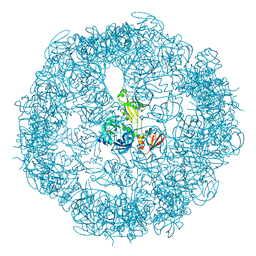 | | re-refined 3CS0 | | 分子名称: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | 著者 | Sauer, R.T, Grant, R.A, Kim, S. | | 登録日 | 2010-09-14 | | 公開日 | 2011-01-19 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QTL
 
 | |
3RSK
 
 | |
3RSD
 
 | |
3RSP
 
 | | STRUCTURE OF THE P93G VARIANT OF RIBONUCLEASE A | | 分子名称: | CHLORIDE ION, RIBONUCLEASE A | | 著者 | Schultz, L.W, Hargraves, S.R, Klink, T.A, Raines, R.T. | | 登録日 | 1997-10-20 | | 公開日 | 1998-04-22 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and stability of the P93G variant of ribonuclease A.
Protein Sci., 7, 1998
|
|
3SD3
 
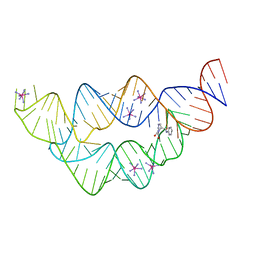 | | The structure of the tetrahydrofolate riboswitch containing a U25C mutation | | 分子名称: | IRIDIUM HEXAMMINE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Tetrahydrofolate riboswitch | | 著者 | Reyes, F.E, Trausch, J.J, Ceres, P, Batey, R.T. | | 登録日 | 2011-06-08 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.
Structure, 19, 2011
|
|
5EA0
 
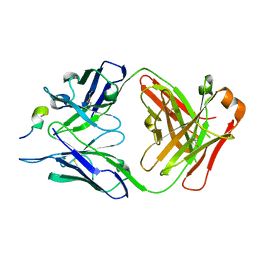 | | Structure of the antibody 7968 with human complement factor H-derived peptide | | 分子名称: | Complement factor H-related protein 2, Heavy chain of antibody 7968 Fab fragment, Light chain of antibody 7968 Fab fragment | | 著者 | Bushey, R.T, Moody, M.A, Nicely, N.I, Alam, S.M, Haynes, B.F, Winkler, M.T, Gottlin, E.B, Campa, M.J, Liao, H.-X, Patz Jr, E.F. | | 登録日 | 2015-10-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Therapeutic Antibody for Cancer, Derived from Single Human B Cells.
Cell Rep, 15, 2016
|
|
5DVB
 
 | |
5EPT
 
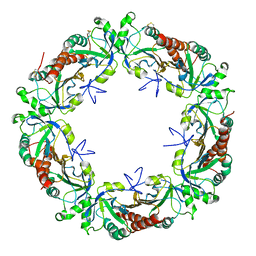 | |
5GM4
 
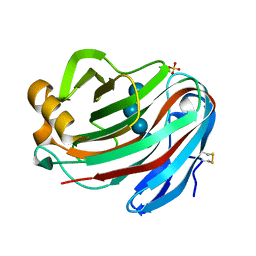 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | 分子名称: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-07-12 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM3
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 | | 分子名称: | CACODYLATE ION, Endoglucanase-1, ZINC ION | | 著者 | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-07-12 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | 分子名称: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-07-13 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
