1XZP
 
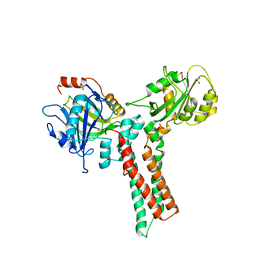 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima | | Descriptor: | Probable tRNA modification GTPase trmE, SULFATE ION | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
1XZQ
 
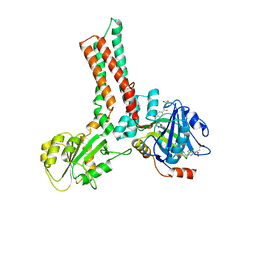 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima complexed with 5-formyl-THF | | Descriptor: | N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, Probable tRNA modification GTPase trmE | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
2GJ8
 
 | |
2GJA
 
 | |
2GJ9
 
 | |
3BRW
 
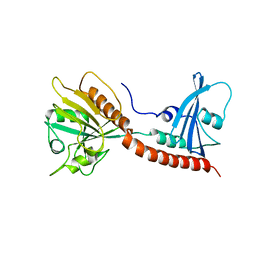 | | Structure of the Rap-RapGAP complex | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scrima, A, Thomas, C, Deaconescu, D, Wittinghofer, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues
Embo J., 27, 2008
|
|
3EI2
 
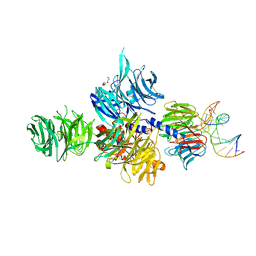 | | Structure of hsDDB1-drDDB2 bound to a 16 bp abasic site containing DNA-duplex | | Descriptor: | 5'-D(*DAP*DAP*DAP*DTP*DGP*DAP*DAP*DTP*(3DR)P*DAP*DAP*DGP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DCP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DTP*DT)-3', DNA damage-binding protein 1, ... | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI1
 
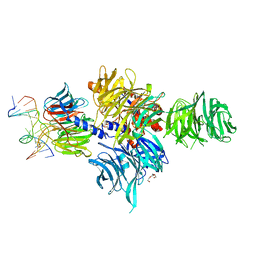 | |
3EI4
 
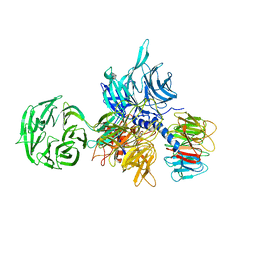 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI3
 
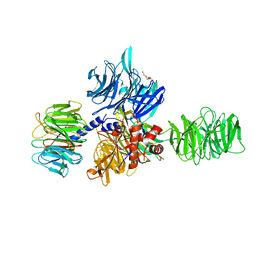 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
4A08
 
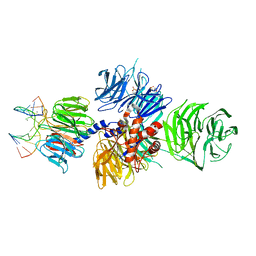 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0A
 
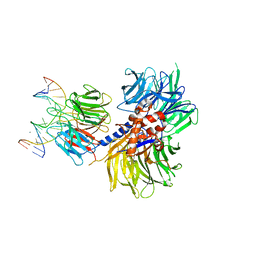 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A09
 
 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0C
 
 | | Structure of the CAND1-CUL4B-RBX1 complex | | Descriptor: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | Authors: | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0B
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
3CP2
 
 | | Crystal structure of GidA from E. coli | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Scrima, A, Meyer, S, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-30 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
5FZP
 
 | | Structure of the dispase autolysis inducing protein from Streptomyces mobaraensis | | Descriptor: | CALCIUM ION, DISPASE AUTOLYSIS-INDUCING PROTEIN, GLYCEROL | | Authors: | Schmelz, S, Fiebig, D, Beck, J, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2016-03-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
J.Biol.Chem., 291, 2016
|
|
6I0I
 
 | |
8RZ6
 
 | | SeMet derivative structure of the condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
2BAP
 
 | |
6XZ5
 
 | |
6GMG
 
 | | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, ... | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase.
FEBS J., 285, 2018
|
|
8QNF
 
 | | Crystal structure of the Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase, FORMIC ACID, GLYCEROL, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
5KE1
 
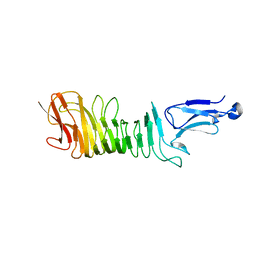 | | Structure of a C-terminal fragment of the IcsA/VirG passenger-domain | | Descriptor: | NICKEL (II) ION, Outer membrane protein IcsA autotransporter | | Authors: | Leupold, S, Scrima, A. | | Deposit date: | 2016-06-09 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the architecture of the Shigella flexneri virulence factor IcsA/VirG and motifs involved in polar distribution and secretion.
J. Struct. Biol., 198, 2017
|
|
