3H1U
 
 | | Structure of ubiquitin in complex with Cd ions | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Qureshi, I.A, Ferron, F, Cheung, P, Lescar, J. | | Deposit date: | 2009-04-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of ubiquitin in complex with cadmium ions
BMC RES NOTES, 2, 2009
|
|
3NI6
 
 | |
4WX1
 
 | |
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
4U8L
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8P
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8N
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8O
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8I
 
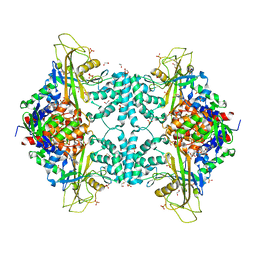 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8K
 
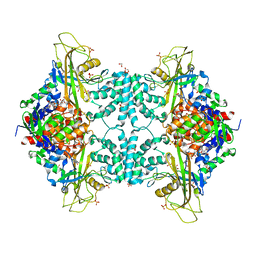 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
6K8Z
 
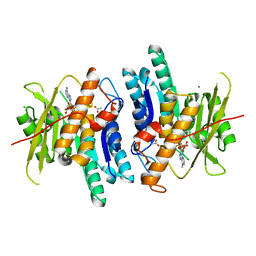 | | Pyridoxal Kinase from Leishmania donovani in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Are, S, Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani.
Int.J.Biol.Macromol., 152, 2020
|
|
6K92
 
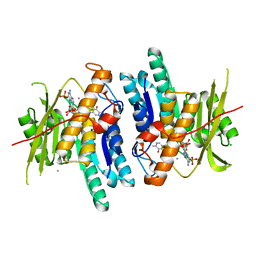 | | Pyridoxal Kinase from Leishmania donovani in complex with ADP and Ginkgotoxin | | Descriptor: | 5-(hydroxymethyl)-4-(methoxymethyl)-2-methylpyridin-3-ol, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Are, S, Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani.
Int.J.Biol.Macromol., 152, 2020
|
|
3LP9
 
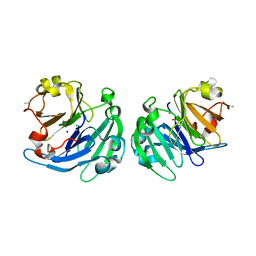 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
5CKU
 
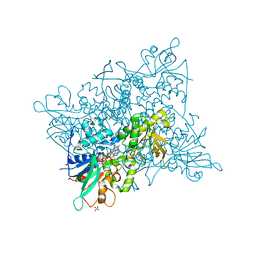 | |
6K91
 
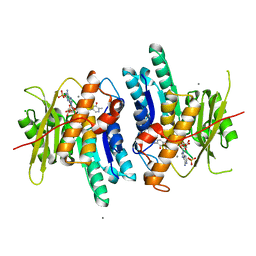 | | Pyridoxal Kinase from Leishmania donovani in complex with ADP and Pyridoxine | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Are, S, Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani.
Int.J.Biol.Macromol., 152, 2020
|
|
6K90
 
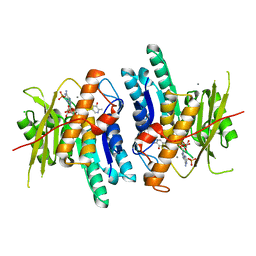 | | Pyridoxal Kinase from Leishmania donovani in complex with ADP and Pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Are, S, Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani.
Int.J.Biol.Macromol., 152, 2020
|
|
6A8C
 
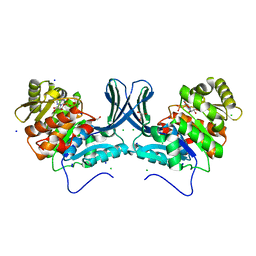 | | Ribokinase from Leishmania donovani with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
5ZWY
 
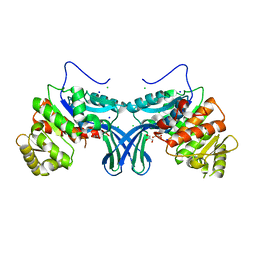 | | Ribokinase from Leishmania donovani | | Descriptor: | CHLORIDE ION, GLYCEROL, Ribokinase, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6A8A
 
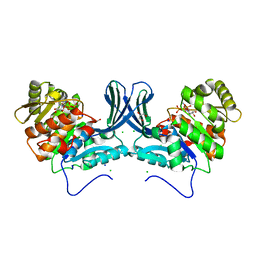 | | Ribokinase from Leishmania donovani with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6A8B
 
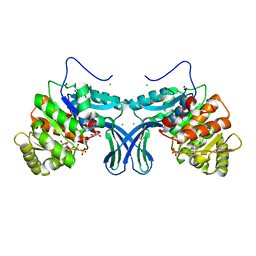 | | Ribokinase from Leishmania donovani with AMPPCP | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Gatreddi, S, Pillalamarri, V, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
3PA7
 
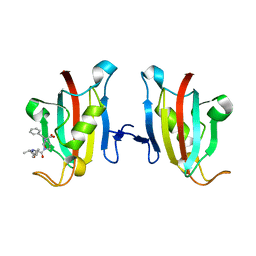 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
7NA0
 
 | |
4ITZ
 
 | |
