2AFN
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE AND A COPPER SITE MUTANT, M150E, THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
1AQ8
 
 | |
1AS8
 
 | |
1AS7
 
 | |
1AS6
 
 | |
1NTD
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE MUTANT M150E THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
1RAQ
 
 | |
1RAP
 
 | |
1YEB
 
 | |
1YEA
 
 | |
3E6S
 
 | |
3E6R
 
 | |
2PP9
 
 | | Nitrate bound wild type oxidized AfNiR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Murphy, M.E.P, Tocheva, E.I. | | Deposit date: | 2007-04-28 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved active site residues limit inhibition of a copper-containing nitrite reductase by small molecules.
Biochemistry, 47, 2008
|
|
3E13
 
 | |
5UJD
 
 | | SbnI from Staphylococcus pseudintermedius | | Descriptor: | FORMIC ACID, Siderophore biosynthesis protein SbnI | | Authors: | Murphy, M.E.P, Verstraete, M.M. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The heme-sensitive regulator SbnI has a bifunctional role in staphyloferrin B production by Staphylococcus aureus .
J.Biol.Chem., 294, 2019
|
|
5UJE
 
 | |
4WCM
 
 | |
5IXG
 
 | | Crystal Structure of Burkholderia cenocepacia BcnB | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, YceI | | Authors: | Loutet, S.A, Murphy, M.E.P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
5IXH
 
 | | Crystal Structure of Burkholderia cenocepacia BcnA | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | Loutet, S.A, Murphy, M.E.P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
6E6I
 
 | | Crystal structure of 4-methyl HOPDA bound to LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | (1Z,3E)-5-carboxy-3-methyl-5-oxo-1-phenylpenta-1,3-dien-1-olate, 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C, Hurst, T.E, Snieckus, V, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2018-07-24 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal- and Serine-Dependent Meta-Cleavage Product Hydrolases Utilize Similar Nucleophile-Activation Strategies
Acs Catalysis, 8, 2018
|
|
8GKD
 
 | |
8GLD
 
 | |
4MP8
 
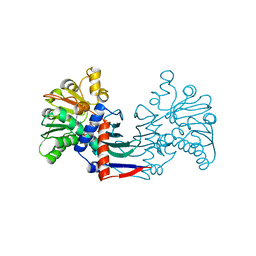 | | Staphyloferrin B precursor biosynthetic enzyme SbnB bound to malonate and NAD+ | | Descriptor: | MALONATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative ornithine cyclodeaminase | | Authors: | Grigg, J.C, Kobylarz, M.J, Rai, D.K, Murphy, M.E.P. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of L-2,3-diaminopropionic Acid, a siderophore and antibiotic precursor.
Chem.Biol., 21, 2014
|
|
4N7K
 
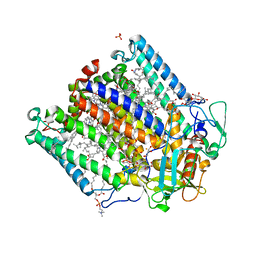 | | Zinc Substituted Reaction Center of the Rhodobacter sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, FE (III) ION, ... | | Authors: | Hardjasa, A, Murphy, M.E.P. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors.
Biochim.Biophys.Acta, 1837, 2013
|
|
4MP3
 
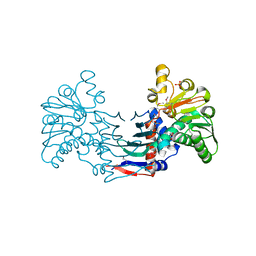 | | Staphyloferrin B precursor biosynthetic enzyme selenomethionine-labeled SbnB | | Descriptor: | GLYCEROL, Putative ornithine cyclodeaminase, SULFATE ION | | Authors: | Grigg, J.C, Kobylarz, M.J, Rai, D.K, Murphy, M.E.P. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Synthesis of L-2,3-diaminopropionic Acid, a siderophore and antibiotic precursor.
Chem.Biol., 21, 2014
|
|
