6AK3
 
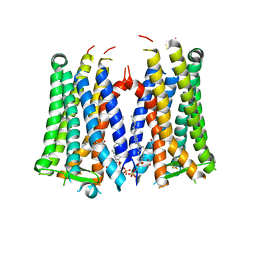 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | Authors: | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-08-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
1X0G
 
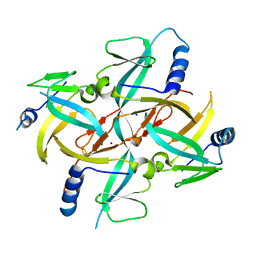 | | Crystal Structure of IscA with the [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IscA, SODIUM ION | | Authors: | Morimoto, K, Yamashita, E, Kondou, Y, Lee, S.J, Tsukihara, T, Nakai, M. | | Deposit date: | 2005-03-22 | | Release date: | 2006-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Asymmetric IscA Homodimer with an Exposed [2Fe-2S] Cluster Suggests the Structural Basis of the Fe-S Cluster Biosynthetic Scaffold.
J.Mol.Biol., 360, 2006
|
|
1ENI
 
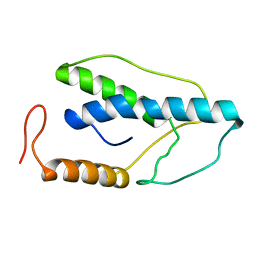 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENK
 
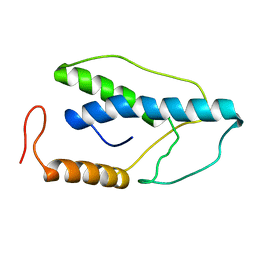 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
3MOO
 
 | | Crystal structure of the HmuO, heme oxygenase from Corynebacterium diphtheriae, in complex with azide-bound verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, AZIDE ION, Heme oxygenase, ... | | Authors: | Omori, K, Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2010-04-22 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Enzymatic ring-opening mechanism of verdoheme by the heme oxygenase: a combined X-ray crystallography and QM/MM study.
J.Am.Chem.Soc., 132, 2010
|
|
3SMV
 
 | | X-ray Crystal Structure of L-Azetidine-2-Carboxylate Hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, GLYCEROL, IMIDAZOLE | | Authors: | Toyoda, M, Mikami, B, Jitsumori, K, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of L-Azetidine-2-carboxylate hydrolase from Pseudomonas sp. strain A2C
To be Published
|
|
2D0O
 
 | | Structure of diol dehydratase-reactivating factor complexed with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
2D0P
 
 | | Structure of diol dehydratase-reactivating factor in nucleotide free form | | Descriptor: | CALCIUM ION, SULFATE ION, diol dehydratase-reactivating factor large subunit, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
7Y51
 
 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis TTE0866 delta100 mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Febs Open Bio, 12, 2022
|
|
6KNB
 
 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6D6S
 
 | |
1CH4
 
 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | Descriptor: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | Deposit date: | 1998-06-11 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
3M0X
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0M
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0V
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0H
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0Y
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329A in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
1POT
 
 | | SPERMIDINE/PUTRESCINE-BINDING PROTEIN COMPLEXED WITH SPERMIDINE (MONOMER FORM) | | Descriptor: | SPERMIDINE, SPERMIDINE/PUTRESCINE-BINDING PROTEIN | | Authors: | Sugiyama, S, Maenaka, K, Matsushima, M, Morikawa, K. | | Deposit date: | 1996-02-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A X-ray structure of the Escherichia coli PotD protein complexed with spermidine and the mechanism of polyamine binding.
Protein Sci., 5, 1996
|
|
3M0L
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
1POY
 
 | | SPERMIDINE/PUTRESCINE-BINDING PROTEIN COMPLEXED WITH SPERMIDINE (DIMER FORM) | | Descriptor: | SPERMIDINE, SPERMIDINE/PUTRESCINE-BINDING PROTEIN | | Authors: | Sugiyama, S, Vassylyev, D.G, Matsushima, M, Morikawa, K. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of PotD, the primary receptor of the polyamine transport system in Escherichia coli.
J.Biol.Chem., 271, 1996
|
|
1A99
 
 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | Descriptor: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | Authors: | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-21 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
1VAS
 
 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
2RN2
 
 | | STRUCTURAL DETAILS OF RIBONUCLEASE H FROM ESCHERICHIA COLI AS REFINED TO AN ATOMIC RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Katayanagi, K, Miyagawa, M, Matsushima, M, Ishikawa, M, Kanaya, S, Nakamura, H, Ikehara, M, Matsuzaki, T, Morikawa, K. | | Deposit date: | 1992-04-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution.
J.Mol.Biol., 223, 1992
|
|
