8ED5
 
 | |
1PV9
 
 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
6C12
 
 | | SDHA-SDHE complex | | Descriptor: | FAD assembly factor SdhE, FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Maher, M.J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of bacterial succinate:quinone oxidoreductase flavoprotein SdhA in complex with its assembly factor SdhE.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6VDA
 
 | |
6VD8
 
 | |
6VD9
 
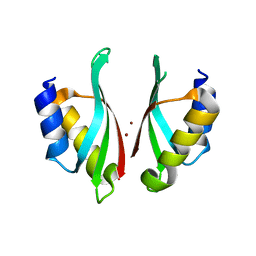 | |
8D2J
 
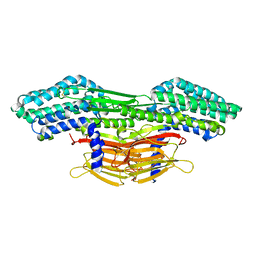 | | A novel insecticidal protein from ferns IPD113_Cow. | | Descriptor: | GLYCEROL, IPD113_Cow | | Authors: | Maher, M.J. | | Deposit date: | 2022-05-30 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel insecticidal proteins from ferns resemble insecticidal proteins from Bacillus thuringiensis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7KR9
 
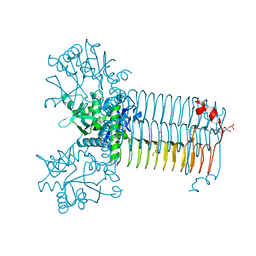 | | Bifunctional enzyme GlmU bound to Zn(II) | | Descriptor: | ACETYL COENZYME *A, Bifunctional protein GlmU, CALCIUM ION, ... | | Authors: | Maher, M.J. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dysregulation of Streptococcus pneumoniae zinc homeostasis breaks ampicillin resistance in a pneumonia infection model.
Cell Rep, 38, 2022
|
|
8ED4
 
 | | Structure of the complex between the arsenite oxidase and its native electron acceptor cytochrome c552 from Pseudorhizobium sp. str. NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AroA, AroB, ... | | Authors: | Maher, M.J, Poddar, N. | | Deposit date: | 2022-09-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the complex between the arsenite oxidase from Pseudorhizobium banfieldiae sp. strain NT-26 and its native electron acceptor cytochrome c 552.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4R98
 
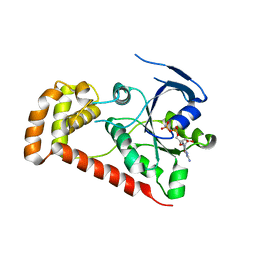 | | Chimera of the N-terminal domain of E. coli FeoB | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ferrous iron transport protein B | | Authors: | Maher, M.J, Jormakka, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A GTPase Chimera Illustrates an Uncoupled Nucleotide Affinity and Release Rate, Providing Insight into the Activation Mechanism.
Biophys.J., 107, 2014
|
|
3OCD
 
 | | Diheme SoxAX - C236M mutant | | Descriptor: | HEME C, SoxA, SoxX | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-09 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
3OA8
 
 | | Diheme SoxAX | | Descriptor: | HEME C, SULFATE ION, SoxA, ... | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
6DU7
 
 | | Glutathione reductase from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, THIOCYANATE ION | | Authors: | Maher, M.J, Sikanyika, M. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The structure and activity of the glutathione reductase from Streptococcus pneumoniae.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6NFR
 
 | | CopC from Pseudomonas fluorescens | | Descriptor: | CopC, SULFATE ION | | Authors: | Maher, M.J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of the CopC protein from Pseudomonas fluorescens reveals amended classifications for the CopC protein family.
J. Inorg. Biochem., 195, 2019
|
|
6NFS
 
 | | CopC from Pseudomonas fluorescens | | Descriptor: | CopC | | Authors: | Maher, M.J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the CopC protein from Pseudomonas fluorescens reveals amended classifications for the CopC protein family.
J. Inorg. Biochem., 195, 2019
|
|
6NFQ
 
 | | CopC from Pseudomonas fluorescens | | Descriptor: | COPPER (II) ION, CopC, YTTRIUM (III) ION | | Authors: | Maher, M.J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the CopC protein from Pseudomonas fluorescens reveals amended classifications for the CopC protein family.
J. Inorg. Biochem., 195, 2019
|
|
3HYR
 
 | |
3HYT
 
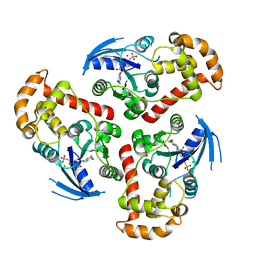 | | Structural Basis of GDP Release and Gating in G Protein Coupled Fe2+ Transport | | Descriptor: | 2-amino-9-(5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-3-O-{[2-(methylamino)phenyl]carbonyl}-beta-D-erythro-pentofuranosyl-2-ulose)-1,9-dihydro-6H-purin-6-one, Ferrous iron transport protein B, MAGNESIUM ION | | Authors: | Maher, M.J, Jormakka, M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of GDP release and gating in G protein coupled Fe(2+) transport.
Embo J., 2009
|
|
6PCE
 
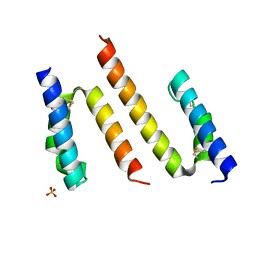 | | Human Coa6 | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog, SULFATE ION | | Authors: | Maher, M.J, Maghool, S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-02 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6.
Life Sci Alliance, 2, 2019
|
|
6PCF
 
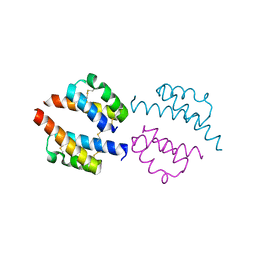 | | Human Coa6: W59C mutant protein | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog | | Authors: | Maher, M.J, Maghool, S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6.
Life Sci Alliance, 2, 2019
|
|
6XAB
 
 | |
6XAD
 
 | |
