1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
2LKQ
 
 | | NMR structure of the lambda 5 22-45 peptide | | Descriptor: | Immunoglobulin lambda-like polypeptide 1 | | Authors: | Elantak, L, Espeli, M, Boned, A, Bornet, O, Breton, C, Feracci, M, Roche, P, Guerlesquin, F, Schiff, C. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation.
J.Biol.Chem., 287, 2012
|
|
2KRB
 
 | | Solution structure of EIF3B-RRM bound to EIF3J peptide | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit J | | Authors: | Elantak, L, Wagner, S, Herrmannova, A, Janoskova, M, Rutkai, E, Lukavsky, P.J, Valasek, L. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The indispensable N-terminal half of eIF3j/HCR1 co-operates with
its structurally conserved binding partner eIF3b/PRT1-RRM and eIF1A in
stringent AUG selection
To be Published
|
|
8EDG
 
 | |
2NLW
 
 | |
8EB5
 
 | |
8RPJ
 
 | | JanthE from Janthinobacterium sp. HH01 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPI
 
 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PP5
 
 | |
8PP2
 
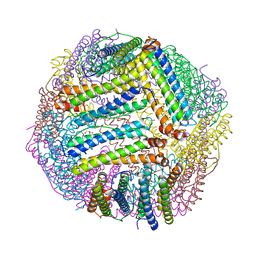 | |
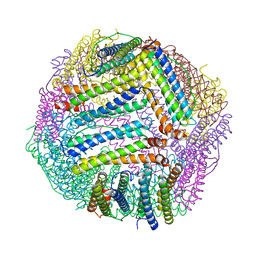
8PP4
 
 | |
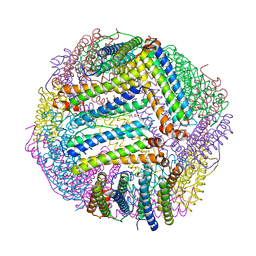
8PP3
 
 | |
8RPH
 
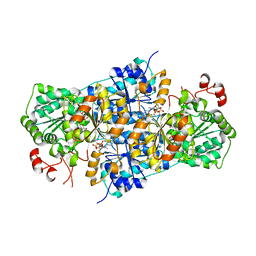 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1Z4H
 
 | | The response regulator TorI belongs to a new family of atypical excisionase | | Descriptor: | Tor inhibition protein | | Authors: | Elantak, L, Ansaldi, M, Guerlesquin, F, Mejean, V, Morelli, X. | | Deposit date: | 2005-03-16 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor
J.Biol.Chem., 280, 2005
|
|
8SJD
 
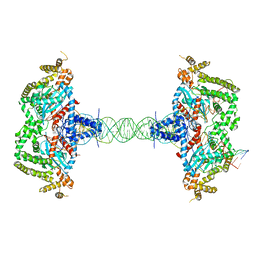 | |
1EMI
 
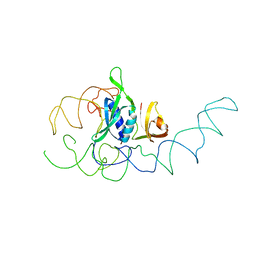 | | STRUCTURE OF 16S RRNA IN THE REGION AROUND RIBOSOMAL PROTEIN S8. | | Descriptor: | 16S RIBOSOMAL RNA, RIBOSOMAL PROTEIN S8 | | Authors: | Lancaster, L, Culver, G.M, Yusupova, G.Z, Cate, J.H, Yuspov, M.M, Noller, H.F. | | Deposit date: | 2000-03-16 | | Release date: | 2000-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | The location of protein S8 and surrounding elements of 16S rRNA in the 70S ribosome from combined use of directed hydroxyl radical probing and X-ray crystallography.
RNA, 6, 2000
|
|
1GX7
 
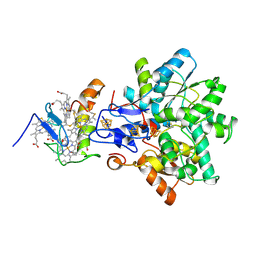 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | Descriptor: | RNA polymerase alpha subunit | | Authors: | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | Deposit date: | 2003-10-06 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
1CNU
 
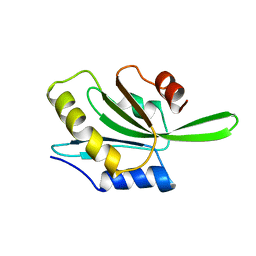 | |
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
6AS9
 
 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 form 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic insights into the genesis of cellular filaments by globular proteins.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
6M5F
 
 | | Crystal structure of HinK, a LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, Y, Lan, L, Cao, Q, Gan, J, Wang, F. | | Deposit date: | 2020-03-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of HinK, a LysR family transcriptional regulator from Pseudomonas aeruginosa
To Be Published
|
|
3STA
 
 | | Crystal structure of ClpP in tetradecameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
