1RPX
 
 | |
8BTT
 
 | | Structure of human RTCB | | Descriptor: | MANGANESE (II) ION, RNA-splicing ligase RtcB homolog | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
8BTX
 
 | | Structure of human Archease | | Descriptor: | Protein archease | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
8ODP
 
 | | Structure of human RTCB with GMPCPP in complex with Archease | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Protein archease, ... | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
8ODO
 
 | | Structure of human guanylylated RTCB in complex with Archease | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
4GMQ
 
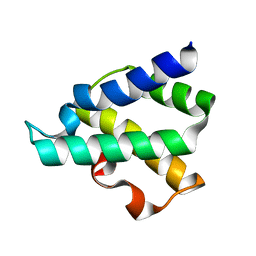 | | Ribosome-binding domain of Zuo1 | | Descriptor: | Putative ribosome associated protein | | Authors: | Kopp, J, Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B9Q
 
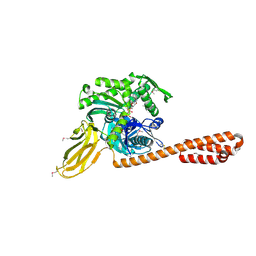 | |
4CIU
 
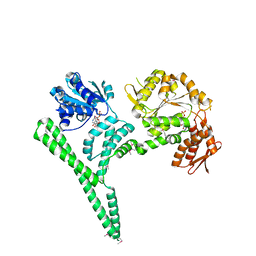 | | Crystal structure of E. coli ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | Authors: | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
5MU7
 
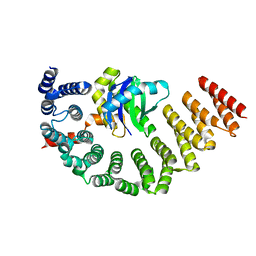 | | Crystal Structure of the beta/delta-COPI Core Complex | | Descriptor: | Coatomer subunit beta, Coatomer subunit delta-like protein | | Authors: | Kopp, J, Aderhold, P, Wieland, F, Sinning, I. | | Deposit date: | 2017-01-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NRO
 
 | | Structure of full-length DnaK with bound J-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaJ, Chaperone protein DnaK, ... | | Authors: | Kopp, J, Kityk, R, Mayer, M.P. | | Deposit date: | 2017-04-25 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones.
Mol. Cell, 69, 2018
|
|
4D73
 
 | | X-ray structure of a peroxiredoxin | | Descriptor: | 1-CYS PEROXIREDOXIN | | Authors: | Staudacher, V, Djuika, C.F, Koduka, J, Schlossarek, S, Kopp, J, Buechler, M, Lanzer, M, Deponte, M. | | Deposit date: | 2014-11-19 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasmodium Falciparum Antioxidant Protein Reveals a Novel Mechanism for Balancing Turnover and Inactivation of Peroxiredoxins
Free Radic.Biol.Med., 85, 2015
|
|
6Z00
 
 | | Arabidopsis thaliana Naa50 in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA-LEU | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6YZZ
 
 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6ZMP
 
 | |
7OJU
 
 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Weidenhausen, J, Kopp, J, Sinning, I. | | Deposit date: | 2021-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
7OVU
 
 | |
7OVV
 
 | | Crystal structure of the Arabidopsis thaliana thialysine acetyltransferase AtNATA2 | | Descriptor: | Probable acetyltransferase NATA1-like, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Layer, D, Kopp, J, Sinning, I. | | Deposit date: | 2021-06-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the Arabidopsis thaliana thialysine acetyltransferase AtNATA2
To Be Published
|
|
6TGX
 
 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with a bisubstrate analogue | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
6TH0
 
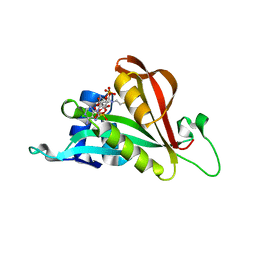 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Acyl-CoA N-acyltransferases (NAT) superfamily protein | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
6SR6
 
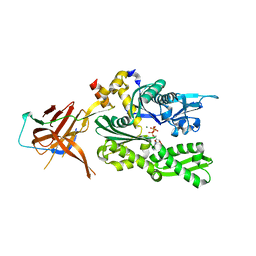 | | Crystal structure of the RAC core with a pseudo substrate bound to Ssz1 SBD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein, ... | | Authors: | Valentin Gese, G, Lapouge, K, Kopp, J, Sinning, I. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb.
Nat Commun, 11, 2020
|
|
3ZRI
 
 | |
4D2U
 
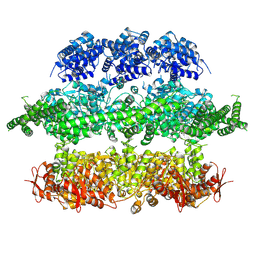 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
3ZRJ
 
 | | Complex of ClpV N-domain with VipB peptide | | Descriptor: | 1,2-ETHANEDIOL, CLPB PROTEIN, VIPB | | Authors: | Lenherr, E.D, Kopp, J, Sinning, I. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Basis for the Unique Role of the Aaa+ Chaperone Clpv in Type Vi Protein Secretion.
J.Biol.Chem., 286, 2011
|
|
4D2Q
 
 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
