5SQY
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5211314110 - (S,S) isomer | | Descriptor: | (1S,2S)-1-{[4-(cyclopropylcarbamamido)-1,3-benzothiazole-7-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SRB
 
 | |
5SQJ
 
 | |
5SQV
 
 | |
6EFJ
 
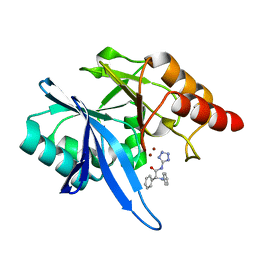 | | Crystal structure of NDM-1 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5SQE
 
 | |
5SR2
 
 | |
5SQN
 
 | |
5SQW
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5014193706 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQS
 
 | |
5SR6
 
 | |
5SQK
 
 | |
5SR1
 
 | |
5SQR
 
 | |
5SQG
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894430 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SR7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4914649782 - (R,R,S) and (S,S,R) isomers | | Descriptor: | (1R,6S,7R)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid, (1S,6R,7S)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SR3
 
 | |
5SQP
 
 | |
5SR4
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z2479779298 - (R,S) and (S,R) isomers | | Descriptor: | (1R,3S)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol, (1S,3R)-3-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclopentan-1-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQF
 
 | |
5SR8
 
 | |
5SQQ
 
 | |
5SR5
 
 | |
5SQT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000833624464 - (R,R) and (S,S) isomers | | Descriptor: | (3R,4R)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid, (3S,4S)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SR9
 
 | |
