6VLO
 
 | | X-ray Structure of the R141 Sugar 4,6-dehydratase from Acanthamoeba polyphaga Minivirus | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative dTDP-D-glucose 4,6-dehydratase, ... | | Authors: | Thoden, J.B, Ferek, J.D, Holden, H.M. | | Deposit date: | 2020-01-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical analysis of a sugar 4,6-dehydratase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
6V33
 
 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
5KGH
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KF9
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGA
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KTC
 
 | | FdhC with bound products: Coenzyme A and 3-[(R)-3-hydroxybutanoylamino]-3,6-dideoxy-d-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
5KTA
 
 | | Apo FdhC- a nucleotide-linked sugar GNAT | | Descriptor: | DIMETHYL SULFOXIDE, FdhC, SULFATE ION | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
5KTD
 
 | | FdhC with bound products: Coenzyme A and dTDP-3-amino-3,6-dideoxy-d-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
1UDB
 
 | |
1UDC
 
 | |
1Z45
 
 | | Crystal structure of the gal10 fusion protein galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae complexed with NAD, UDP-glucose, and galactose | | Descriptor: | GAL10 bifunctional protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2005-03-15 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The molecular architecture of galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae.
J.Biol.Chem., 280, 2005
|
|
1UDA
 
 | |
2PAM
 
 | | Structure of a H49N, H51N double mutant dTDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase from Aneurinibacillus thermoaerophilus complexed with TDP | | Descriptor: | DTDP-6-deoxy-3,4-keto-hexulose isomerase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Davis, M.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray Structure of dTDP-4-Keto-6-deoxy-D-glucose-3,4-ketoisomerase.
J.Biol.Chem., 282, 2007
|
|
2PAE
 
 | | Structure of a H49N mutant dTDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase from Aneurinibacillus thermoaerophilus in complex with TDP | | Descriptor: | DTDP-6-deoxy-3,4-keto-hexulose isomerase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Davis, M.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray Structure of dTDP-4-Keto-6-deoxy-D-glucose-3,4-ketoisomerase.
J.Biol.Chem., 282, 2007
|
|
2A2C
 
 | |
2AQO
 
 | | Crystal structure of E. coli Isoaspartyl Dipeptidase Mutant E77Q | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
1WUU
 
 | | crystal structure of human galactokinase complexed with MgAMPPNP and galactose | | Descriptor: | Galactokinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Thoden, J.B, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Structure of Human Galactokinase: IMPLICATIONS FOR TYPE II GALACTOSEMIA
J.Biol.Chem., 280, 2005
|
|
2AJ4
 
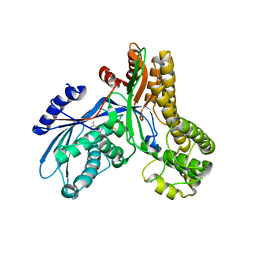 | | Crystal structure of Saccharomyces cerevisiae Galactokinase in complex with galactose and Mg:AMPPNP | | Descriptor: | CHLORIDE ION, Galactokinase, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Sellick, C.A, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer
J.Biol.Chem., 280, 2005
|
|
1WVC
 
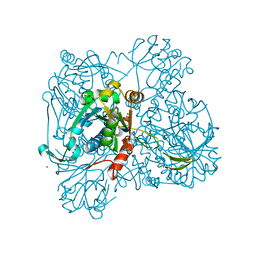 | | alpha-D-glucose-1-phosphate cytidylyltransferase complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Glucose-1-phosphate cytidylyltransferase, MAGNESIUM ION, ... | | Authors: | Koropatkin, N.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2004-12-14 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of alpha-D-Glucose-1-phosphate cytidylyltransferase from Salmonella typhi.
J.Biol.Chem., 280, 2005
|
|
3VBL
 
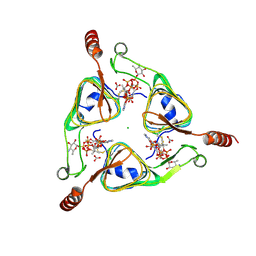 | | Crystal Structure of the S84C mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
1WVG
 
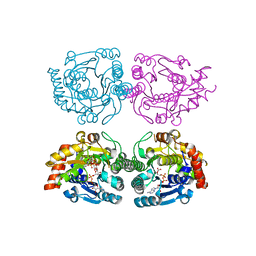 | | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CDP-glucose 4,6-dehydratase, CYTIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE | | Authors: | Koropatkin, N.M, Holden, H.M. | | Deposit date: | 2004-12-15 | | Release date: | 2005-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi complexed with CDP-D-xylose.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3VBK
 
 | | Crystal Structure of the S84A mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
3VBM
 
 | |
3VBP
 
 | |
