3NBS
 
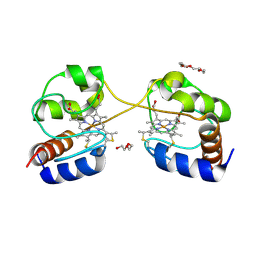 | | Crystal structure of dimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBT
 
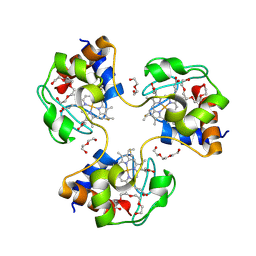 | | Crystal structure of trimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5GQC
 
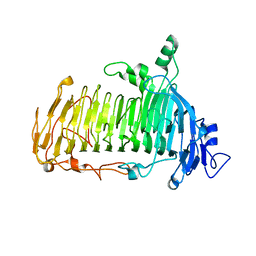 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQG
 
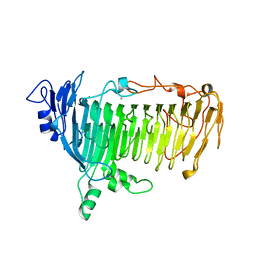 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQF
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
3ACT
 
 | |
3ABZ
 
 | | Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus | | Descriptor: | Beta-glucosidase I, GLYCEROL | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
3ACS
 
 | |
3AFJ
 
 | |
4ZLF
 
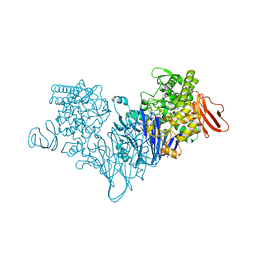 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLI
 
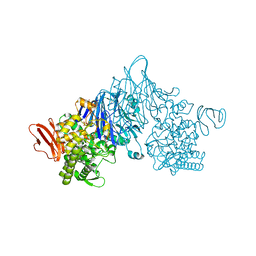 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
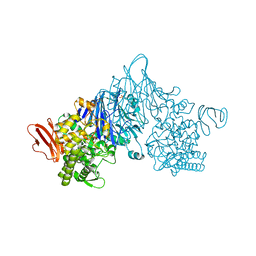 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLG
 
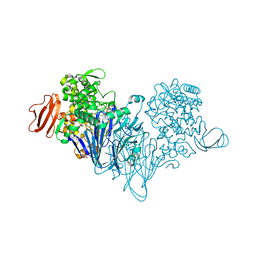 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZN
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
3AC0
 
 | | Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose | | Descriptor: | Beta-glucosidase I, beta-D-glucopyranose | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
7V1W
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-arabinofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase (alphaFFase1), beta-D-arabinofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1X
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-fructofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase, beta-D-fructofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
8I5L
 
 | |
6HUS
 
 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
6HUR
 
 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 2'-fucosyllactose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Katayama, T, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
4JAW
 
 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
