1RI5
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI4
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI2
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1J5D
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803-MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
1J5C
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
1YN9
 
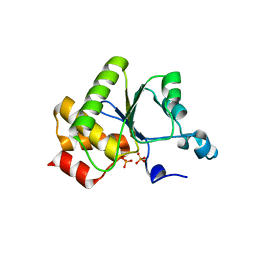 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
1K9E
 
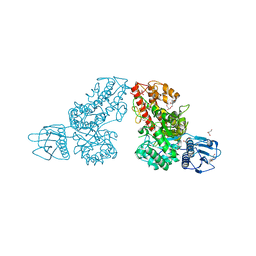 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9D
 
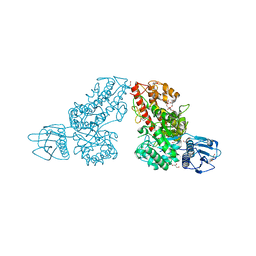 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
3BGY
 
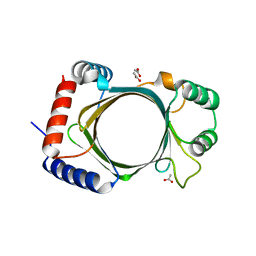 | |
1LY1
 
 | |
1L8N
 
 | | The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose | | Descriptor: | 4-O-methyl-beta-D-glucopyranuronic acid, ALPHA-D-GLUCURONIDASE, GLYCEROL, ... | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-03-21 | | Release date: | 2003-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1MQR
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE (E386Q) FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MQQ
 
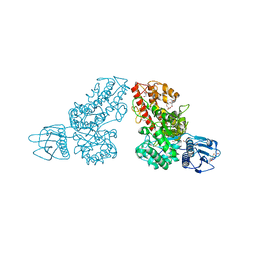 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL, alpha-D-glucopyranuronic acid | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1K9F
 
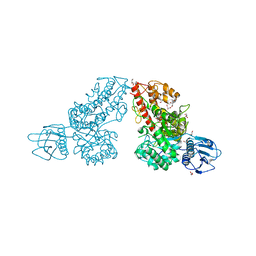 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
3P43
 
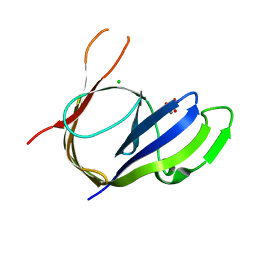 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
3OQ2
 
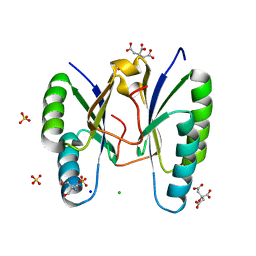 | | Structure of a CRISPR associated protein Cas2 from Desulfovibrio vulgaris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Samai, P, Smith, P, Shuman, S. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3P4H
 
 | | Structures of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Zhu, H, Shuman, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
2IA5
 
 | | T4 polynucleotide kinase/phosphatase with bound sulfate and magnesium. | | Descriptor: | ARSENIC, MAGNESIUM ION, Polynucleotide kinase, ... | | Authors: | Zhu, H, Smith, P.C, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2006-09-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis of the 3' phosphatase component of T4 polynucleotide kinase/phosphatase.
Virology, 366, 2007
|
|
3PQV
 
 | | Cyclase homolog | | Descriptor: | D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tanaka, N, Smith, P, Shuman, S. | | Deposit date: | 2010-11-27 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis.
Rna, 17, 2011
|
|
5V9X
 
 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | Descriptor: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | Deposit date: | 2017-03-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
1FVI
 
 | | CRYSTAL STRUCTURE OF CHLORELLA VIRUS DNA LIGASE-ADENYLATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORELLA VIRUS DNA LIGASE-ADENYLATE, SULFATE ION | | Authors: | Odell, M, Sriskanda, V, Shuman, S, Nikolov, D.B. | | Deposit date: | 2000-09-20 | | Release date: | 2000-11-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining.
Mol.Cell, 6, 2000
|
|
1I9S
 
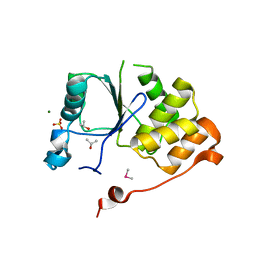 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
1I9T
 
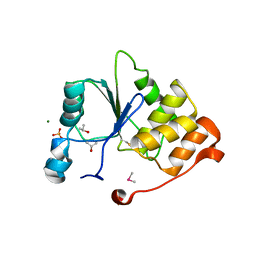 | | CRYSTAL STRUCTURE OF THE OXIDIZED RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
2LJ6
 
 | |
