6S2B
 
 | | Structure of beta-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosyl-erythritol | | Descriptor: | (2~{S},3~{R})-4-[(2~{R},3~{S},4~{S},5~{R})-2,5-bis(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxybutane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New insights into the molecular mechanism behind mannitol and erythritol fructosylation by beta-fructofuranosidase from Schwanniomyces occidentalis.
Sci Rep, 11, 2021
|
|
6S1T
 
 | |
6SXP
 
 | |
6SXY
 
 | |
6SYA
 
 | |
6SYL
 
 | |
1B2R
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
1E64
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LYS 75 REPLACED BY GLN (K75Q) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1E62
 
 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Arg (K75R) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1W87
 
 | |
8BBI
 
 | |
2BSA
 
 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
6R5V
 
 | |
6R5P
 
 | |
6R5U
 
 | |
6R5T
 
 | |
6R5N
 
 | |
6R5O
 
 | |
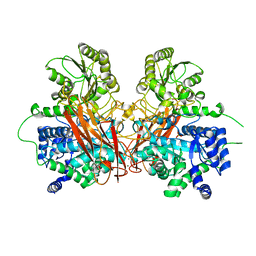
6R5I
 
 | |
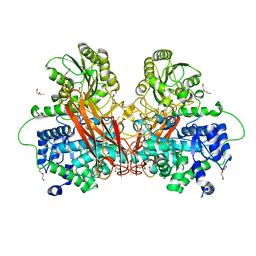
6R5R
 
 | |
3URJ
 
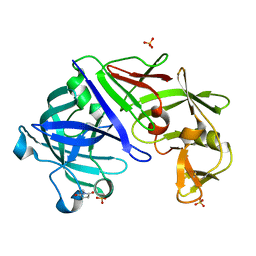 | | Type IV native endothiapepsin | | Descriptor: | Endothiapepsin, SULFATE ION | | Authors: | Bailey, D, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UTL
 
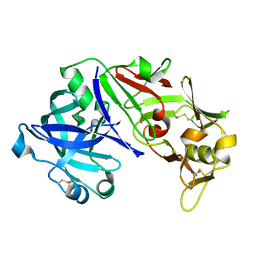 | | Human pepsin 3b | | Descriptor: | Pepsin A | | Authors: | Coker, A, Cooper, J.B. | | Deposit date: | 2011-11-25 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1GR1
 
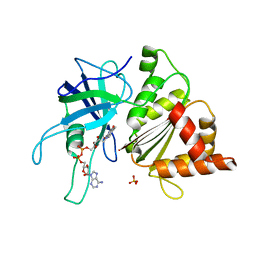 | | Structure of Ferredoxin-NADP+ Reductase with Glu 139 replaced by Lys (E139K) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Role of Glutamic Acid 139 of Anabena Ferrodoxin-Nadp+ Reductase in the Interaction with Substrates
Eur.J.Biochem., 269, 2002
|
|
7AUU
 
 | |
7AUO
 
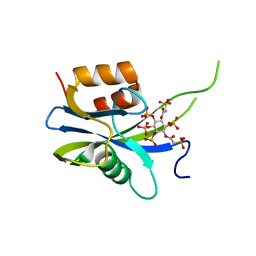 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PA-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(2-oxoethane-2,1-diyl)]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
