2AMV
 
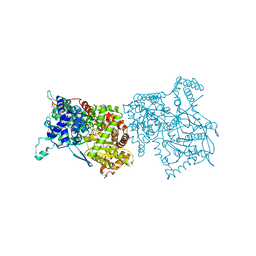 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
6PLI
 
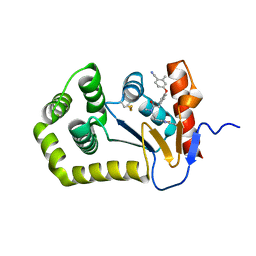 | | Crystal Structure of EcDsbA in a complex with purified oxadiazole 11 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-methyl-N-[2-(5-methyl-1,2,4-oxadiazol-3-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-07-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PC9
 
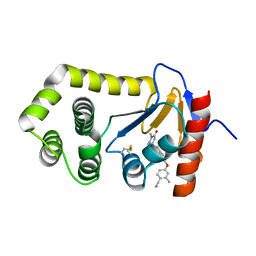 | | Crystal Structure of EcDsbA in a complex with purified methylpiperazinone 6 | | Descriptor: | 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
6PIQ
 
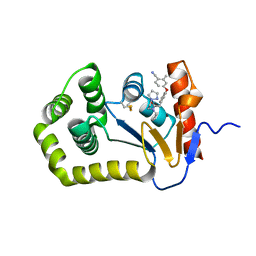 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product G6 (pyrazole 9) | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG2
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PBI
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG1
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product F1 (methylpiperazinone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PGJ
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PDH
 
 | | Crystal Structure of EcDsbA in a complex with purified pyrazole 9 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-19 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
4WET
 
 | | Crystal structure of E.Coli DsbA in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-tyrosine, SODIUM ION, ... | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6BQX
 
 | | Crystal structure of Escherichia coli DsbA in complex with N-methyl-1-(4-phenoxyphenyl)methanamine | | Descriptor: | N-methyl-1-(4-phenoxyphenyl)methanamine, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
2JKZ
 
 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (ORTHORHOMBIC CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
2JKY
 
 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (TETRAGONAL CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
1BL0
 
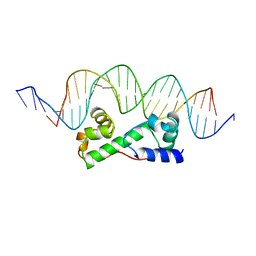 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | Authors: | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | Deposit date: | 1998-07-22 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8GPB
 
 | |
1BQ7
 
 | |
4K2D
 
 | | Crystal structure of Burkholderia Pseudomallei DsbA | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein | | Authors: | McMahon, R.M. | | Deposit date: | 2013-04-09 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disarming Burkholderia pseudomallei: Structural and Functional Characterization of a Disulfide Oxidoreductase (DsbA) Required for Virulence In Vivo.
Antioxid Redox Signal, 20, 2014
|
|
3H93
 
 | | Crystal Structure of Pseudomonas aeruginosa DsbA | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein dsbA | | Authors: | Shouldice, S.R. | | Deposit date: | 2009-04-29 | | Release date: | 2009-12-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization of the DsbA Oxidative Folding Catalyst from Pseudomonas aeruginosa Reveals a Highly Oxidizing Protein that Binds Small Molecules.
Antioxid Redox Signal, 12, 2010
|
|
3HYP
 
 | |
3HXS
 
 | | Crystal Structure of Bacteroides fragilis TrxP | | Descriptor: | Thioredoxin, ZINC ION | | Authors: | Shouldice, S.R. | | Deposit date: | 2009-06-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | In vivo oxidative protein folding can be facilitated by oxidation-reduction cycling
Mol.Microbiol., 75, 2010
|
|
9GPB
 
 | |
2GPN
 
 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
6GPB
 
 | |
1GPY
 
 | |
