3WR3
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WKU
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR8
 
 | | Crystal structure of DesB from Sphingobium sp. strain SYK-6 | | Descriptor: | FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
5XNY
 
 | | Crystal structure of CreD | | Descriptor: | CreD | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XNZ
 
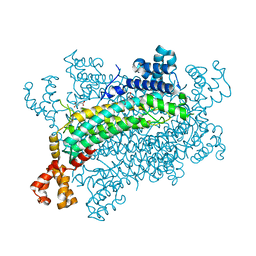 | | Crystal structure of CreD complex with fumarate | | Descriptor: | CreD, FUMARIC ACID | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
1B4U
 
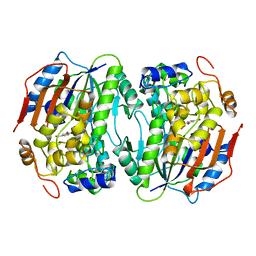 | | PROTOCATECHUATE 4,5-DIOXYGENASE (LIGAB) IN COMPLEX WITH PROTOCATECHUATE (PCA) | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 4,5-DIOXYGENASE | | Authors: | Sugimoto, K, Senda, T, Mitsui, Y. | | Deposit date: | 1998-12-29 | | Release date: | 1999-08-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure Fold.Des., 7, 1999
|
|
5ZM4
 
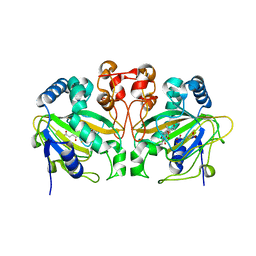 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AndA with preandiloid C | | Descriptor: | (6aS,8aR,12aS,12bR,13aR)-5,6a,9,9,12a,13a-hexamethyl-7,8,8a,9,12a,12b,13,13a-octahydro-3H-benzo[a]furo[3,4-j]xanthene-3,4,10(1H,6aH)-trione, 2-OXOGLUTARIC ACID, Dioxygenase andA, ... | | Authors: | Nakashima, Y, Senda, T. | | Deposit date: | 2018-04-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Computational Bases for Dramatic Skeletal Rearrangement in Anditomin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
5ZM2
 
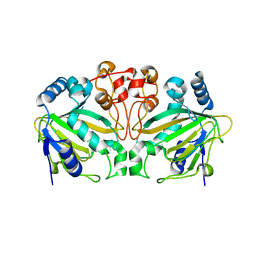 | |
5GGI
 
 | | Crystal structure of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Mn, UDP and Mannosyl-peptide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGK
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-mannopyranoside, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGO
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGF
 
 | |
5GGJ
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-alpha-pNP | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGL
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-alpha-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGP
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta1,2-Man-peptide | | Descriptor: | 10-mer Peptide from Dystroglycan, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGG
 
 | |
5GGN
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1UI5
 
 | |
1UI6
 
 | |
1KW3
 
 | | Crystal structure of 2,3-dihydroxybiphenyal dioxygenase (BphC) at 1.45 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-Dihydroxybiphenyl dioxygenase, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KWC
 
 | | The His145Ala mutant of 2,3-dihydroxybiphenyl dioxygenase in complex with 2,3-dihydroxybiphenyl | | Descriptor: | 2,3-dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW9
 
 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 2.0A resolution | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KWB
 
 | | Crystal structure of the His145Ala mutant of 2,3-dihydroxybipheny dioxygenase (BphC) | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW8
 
 | | Crystal structure of BphC-2,3-dihydroxybiphenyl-NO complex | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW6
 
 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 1.45 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
