3GNL
 
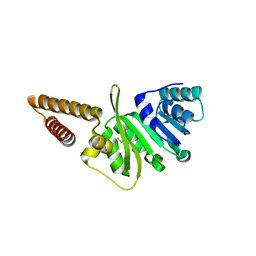 | | Structure of uncharacterized protein (LMOf2365_1472) from Listeria monocytogenes serotype 4b | | Descriptor: | uncharacterized protein, DUF633, LMOf2365_1472 | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-17 | | Release date: | 2009-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of uncharacterized protein (LMOf2365_1472) from
Listeria monocytogenes serotype 4b
To be Published
|
|
3GP4
 
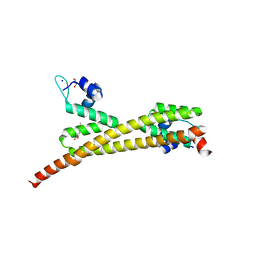 | | Crystal structure of putative MerR family transcriptional regulator from listeria monocytogenes | | Descriptor: | D-METHIONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of putative MerR family transcriptional regulator from Listeria monocytogenes
To be published
|
|
3E18
 
 | | CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-02 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua
To be Published
|
|
3E8V
 
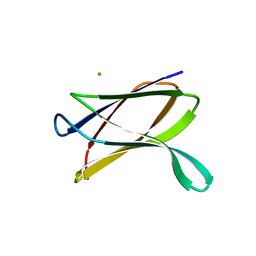 | | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis | | Descriptor: | Possible transglutaminase-family protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis
To be Published
|
|
3ED4
 
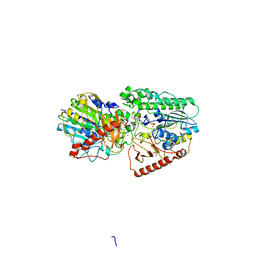 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
3EOI
 
 | | CRYSTAL STRUCTURE OF putative PROTEIN PilM from Escherichia coli B7A | | Descriptor: | PilM | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J.B, Sauder, J.M, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3EEY
 
 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
3DTN
 
 | | Crystal structure of putative Methyltransferase-MM_2633 from Methanosarcina mazei . | | Descriptor: | ACETATE ION, CALCIUM ION, Putative Methyltransferase MM_2633 | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of putative Methyltransferase-MM_2633 from Methanosarcina mazei
To be published
|
|
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | Descriptor: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
3DTC
 
 | | Crystal structure of mixed-lineage kinase MLK1 complexed with compound 16 | | Descriptor: | 12-(2-hydroxyethyl)-2-(1-methylethoxy)-13,14-dihydronaphtho[2,1-a]pyrrolo[3,4-c]carbazol-5(12H)-one, Mitogen-activated protein kinase kinase kinase 9, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2008-07-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-lineage kinase 1 crystallography, and oral in vivo activity in 1-methyl-4-phenyltetrahydropyridine models.
J.Med.Chem., 51, 2008
|
|
3GTZ
 
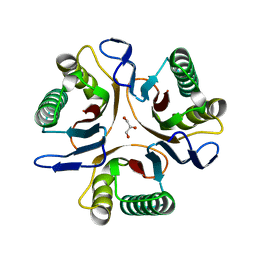 | | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium | | Descriptor: | GLYCEROL, Putative translation initiation inhibitor | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-28 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium
To be Published
|
|
3GMF
 
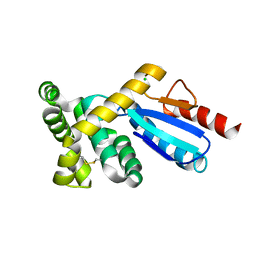 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3GT5
 
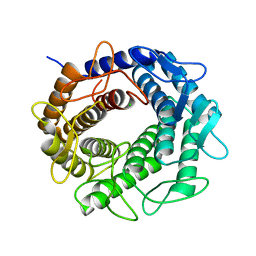 | | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa | | Descriptor: | CHLORIDE ION, N-acetylglucosamine 2-epimerase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa
To be Published
|
|
3H7V
 
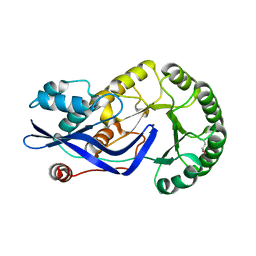 | | CRYSTAL STRUCTURE OF O-SUCCINYLBENZOATE SYNTHASE FROM THERMOSYNECHOCOCCUS ELONGATUS BP-1 complexed with MG in the active site | | Descriptor: | MAGNESIUM ION, O-SUCCINYLBENZOATE SYNTHASE | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ES7
 
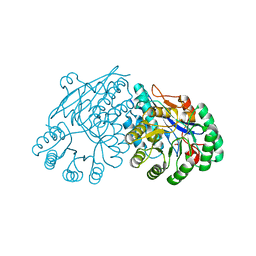 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3EVN
 
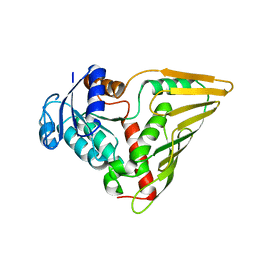 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Streptococcus agalactiae 2603V/r | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative Gfo/Idh/MocA family oxidoreductase from Streptococcus agalactiae
2603V/r
To be Published
|
|
3EZY
 
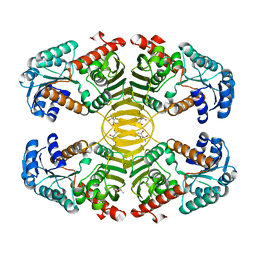 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
3EWC
 
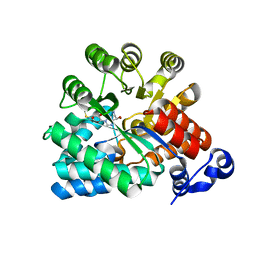 | | Crystal Structure of adenosine deaminase from Plasmodial vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
3EYE
 
 | | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli | | Descriptor: | PTS system N-acetylgalactosamine-specific IIB component 1 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli
To be Published
|
|
3EVT
 
 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
3F5Q
 
 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3FON
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3FDU
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase/isomerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii
To be Published
|
|
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3H5I
 
 | | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans | | Descriptor: | CHLORIDE ION, Response regulator/sensory box protein/GGDEF domain protein, SODIUM ION | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans
To be Published
|
|
