3DWV
 
 | |
4D2U
 
 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2Q
 
 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
3DEF
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
3FEM
 
 | |
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
5CK4
 
 | |
5CK5
 
 | |
5CK3
 
 | | Signal recognition particle receptor SRb-GTP/SRX complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jadhav, B.R, Wild, K, Sinning, I. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Structure, 23, 2015
|
|
5E4X
 
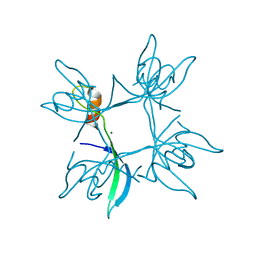 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
5E4W
 
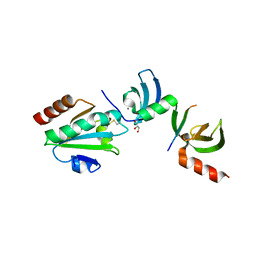 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
6YGU
 
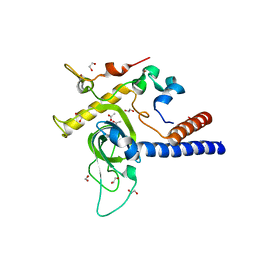 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
6YFV
 
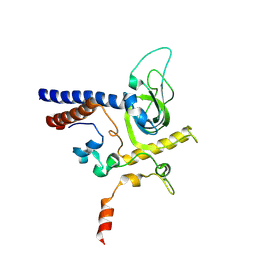 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
5EM2
 
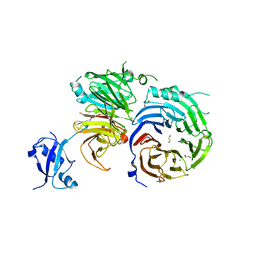 | | Crystal structure of the Erb1-Ytm1 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | Authors: | Ahmed, Y.L, Sinning, I. | | Deposit date: | 2015-11-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
6Z00
 
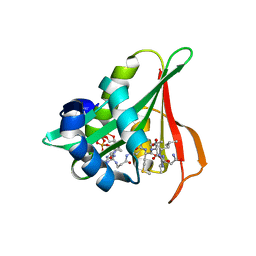 | | Arabidopsis thaliana Naa50 in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA-LEU | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6YZZ
 
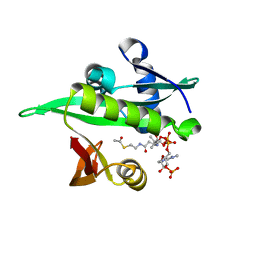 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
5BY8
 
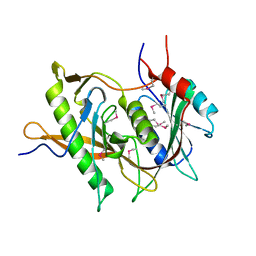 | | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis | | Descriptor: | Rpf2, Rrs1 | | Authors: | Kharde, S, Calvino, F.R, Gumiero, A, Wild, K, Sinning, I. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
3DEO
 
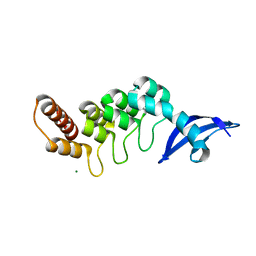 | |
6TGX
 
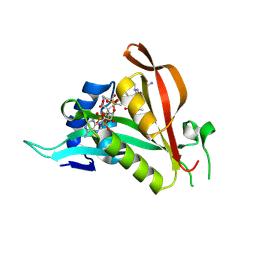 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with a bisubstrate analogue | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
6TH0
 
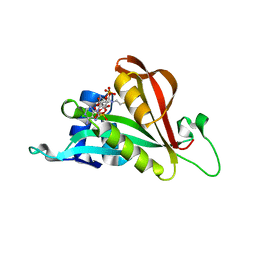 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Acyl-CoA N-acyltransferases (NAT) superfamily protein | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
5L3W
 
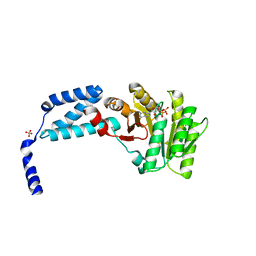 | | Structure of the crenarchaeal FtsY GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle receptor FtsY | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3R
 
 | | Structure of the GTPase heterodimer of chloroplast SRP54 and FtsY from Arabidopsis thaliana | | Descriptor: | Cell division protein FtsY homolog, chloroplastic, GLYCEROL, ... | | Authors: | Bange, G, Kribelbauer, J, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3Q
 
 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wild, K, Segnitz, B, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3V
 
 | | Structure of the crenarchaeal SRP54 GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle 54 kDa protein | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
