1KG5
 
 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
6F1F
 
 | | The methylene thioacetal BPTI (Bovine Pancreatic Trypsin Inhibitor) mutant structure | | Descriptor: | GLYCEROL, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Lansky, S, Mousa, R, Metanis, N, Shoham, G. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.716 Å) | | Cite: | BPTI folding revisited: switching a disulfide into methylene thioacetal reveals a previously hidden path.
Chem Sci, 9, 2018
|
|
6H3M
 
 | |
1K3X
 
 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1K82
 
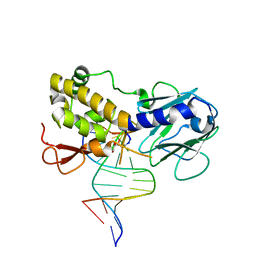 | | Crystal structure of E.coli formamidopyrimidine-DNA glycosylase (Fpg) covalently trapped with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*CP*TP*CP*CP*TP*GP*G)-3', ZINC ION, ... | | Authors: | Gilboa, R, Zharkov, D.O, Golan, G, Fernandes, A.S, Gerchman, S.E, Matz, E, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA.
J.Biol.Chem., 277, 2002
|
|
1ARL
 
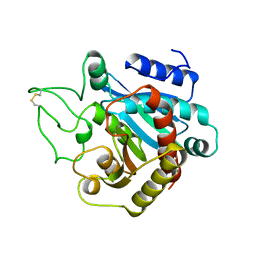 | | CARBOXYPEPTIDASE A WITH ZN REMOVED | | Descriptor: | APO-CARBOXYPEPTIDASE A=ALPHA= (COX) | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-01 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1K3W
 
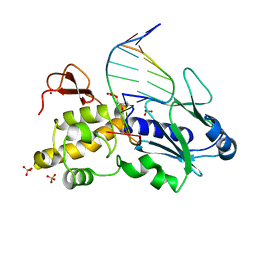 | | Crystal structure of a trapped reaction intermediate of the DNA Repair Enzyme Endonuclease VIII with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1ARM
 
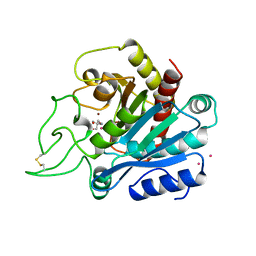 | | CARBOXYPEPTIDASE A WITH ZN REPLACED BY HG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, HG-CARBOXYPEPTIDASE A=ALPHA= (COX), ... | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-17 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1EA7
 
 | | Sphericase | | Descriptor: | CALCIUM ION, SERINE PROTEASE | | Authors: | Almog, O, Gonzalez, A, Klein, D, Braun, S, Shoham, G. | | Deposit date: | 2001-07-10 | | Release date: | 2002-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The 0.93A Crystal Structure of Sphericase: A Calcium-Loaded Serine Protease from Bacillus Sphaericus
J.Mol.Biol., 332, 2003
|
|
2EXJ
 
 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2BS9
 
 | | Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus | | Descriptor: | BETA-XYLOSIDASE, CALCIUM ION | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
2BFG
 
 | | crystal structure of beta-xylosidase (fam GH39) in complex with dinitrophenyl-beta-xyloside and covalently bound xyloside | | Descriptor: | 2,5-DINITROPHENOL, BETA-XYLOSIDASE, SODIUM ION, ... | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
2EXI
 
 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXH
 
 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2IXT
 
 | | SPHERICASE | | Descriptor: | 36KDA PROTEASE, CALCIUM ION | | Authors: | Almog, O, Gonzalez, A, Godin, N. | | Deposit date: | 2006-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | The Crystal Structures of the Psychrophilic Subtilisin S41 and the Mesophilic Subtilisin Sph Reveal the Same Calcium-Loaded State.
Proteins, 74, 2009
|
|
2OQ4
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*C*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*G*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Grollman, A.P, Shoahm, G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
3D43
 
 | | The crystal structure of Sph at 0.8A | | Descriptor: | CALCIUM ION, Sphericase | | Authors: | Almog, O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
2GKO
 
 | | S41 Psychrophilic Protease | | Descriptor: | CALCIUM ION, SODIUM ION, microbial serine proteinases; subtilisin, ... | | Authors: | Walter, R.L, Mekel, M.J, Grayling, R.A, Arnold, F.H, Wintrode, P.L, Almog, O. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
1EE3
 
 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELL
 
 | | CADMIUM-SUBSTITUTED BOVINE PANCREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 7.5 AND 0.25 M CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELM
 
 | | CADMIUM-SUBSTITUTED BOVINE PACREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 5.5 AND 2 MM CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
