8KDA
 
 | | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, MAGNESIUM ION, RNA-free ribonuclease P | | Authors: | Teramoto, T, Adachi, N, Yokogawa, T, Koyasu, T, Mayanagi, K, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis of transfer RNA processing by bacterial minimal RNase P
Nat Commun, 16, 2025
|
|
8KD9
 
 | | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Mayanagi, K, Yokogawa, T, Adachi, N, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis of transfer RNA processing by bacterial minimal RNase P
Nat Commun, 16, 2025
|
|
8HH0
 
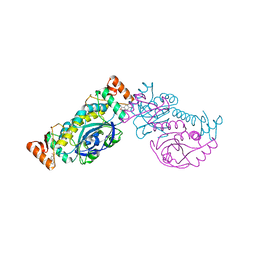 | |
8HLA
 
 | |
5XDH
 
 | | His/DOPA ligated cytochrome c from an anammox organism KSU-1 | | Descriptor: | ACETATE ION, HEME C, Putative cytochrome c, ... | | Authors: | Hira, D, Kitamura, R, Nakamura, T, Yamagata, Y, Furukawa, K, Fujii, T. | | Deposit date: | 2017-03-28 | | Release date: | 2018-03-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Anammox Organism KSU-1 Expresses a Novel His/DOPA Ligated Cytochrome c.
J. Mol. Biol., 430, 2018
|
|
7X1L
 
 | | Malate dehydrogenase from Geobacillus stearothermophilus (gs-MDH) delta E311 mutant complexed with Nicotinamide Adenine Dinucleotide (NAD+) | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Reducing substrate inhibition of malate dehydrogenase from Geobacillus stearothermophilus by C-terminal truncation.
Protein Eng.Des.Sel., 35, 2022
|
|
5HBP
 
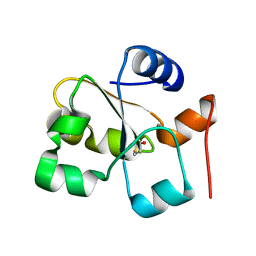 | | The crystal of rhodanese domain of YgaP treated with SNOC | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBQ
 
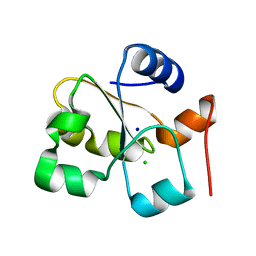 | | C63D mutant of the rhodanese domain of YgaP | | Descriptor: | CHLORIDE ION, Inner membrane protein YgaP, SODIUM ION | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-02 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBO
 
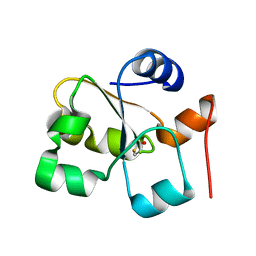 | | Native rhodanese domain of YgaP prepared without DDT is both S-nitrosylated and S-sulfhydrated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBL
 
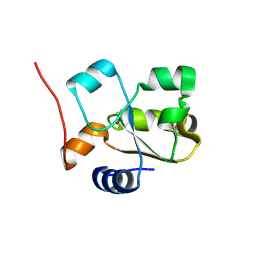 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
3WE7
 
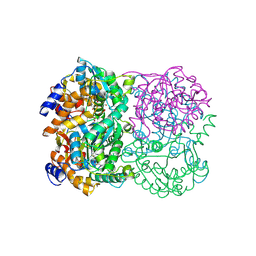 | | Crystal Structure of Diacetylchitobiose Deacetylase from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Mine, S, Nakamura, T, Fukuda, Y, Inoue, T, Uegaki, K, Sato, T. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
7Y51
 
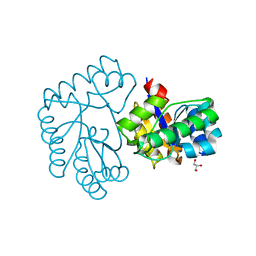 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis TTE0866 delta100 mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Febs Open Bio, 12, 2022
|
|
3VP8
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p | | Descriptor: | General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3WUC
 
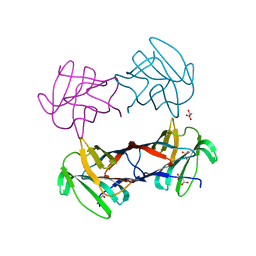 | | X-ray crystal structure of Xenopus laevis galectin-Va | | Descriptor: | Galectin, MALONIC ACID, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
3WUD
 
 | | X-ray crystal structure of Xenopus laevis galectin-Ib | | Descriptor: | Galectin, SULFATE ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
6L0R
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | Descriptor: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
7FBW
 
 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis | | Descriptor: | NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nishiya, Y, Nakamura, T. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6KRP
 
 | |
6KRQ
 
 | |
6KRK
 
 | |
6KRR
 
 | |
6KRM
 
 | |
7BY9
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) complexed with Oxaloacetic Acid (OAA) and Nicotinamide Adenine Dinucleotide (NAD) | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | Authors: | Shimozawa, Y, Nakamura, T, Himiyama, T, Nishiya, Y. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis and reaction mechanism of malate dehydrogenase from Geobacillus stearothermophilus.
J.Biochem., 170, 2021
|
|
7BY8
 
 | |
