4F0X
 
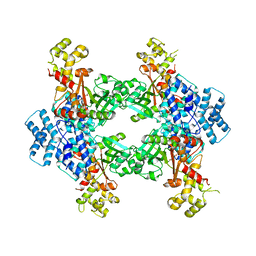 | | Crystal structure of human Malonyl-CoA Decarboxylase (Peroxisomal Isoform) | | Descriptor: | Malonyl-CoA decarboxylase, mitochondrial, N~3~-[(2R)-2-hydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-3,3-dimethylbutanoyl]-beta-alaninamide | | Authors: | Aparicio, D, Perez, R, Fita, I. | | Deposit date: | 2012-05-05 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Asymmetry and Disulfide Bridges among Subunits Modulate the Activity of Human Malonyl-CoA Decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4FGW
 
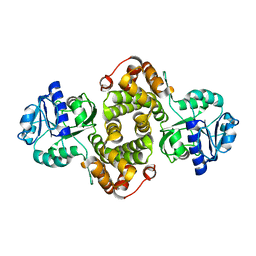 | | Structure of Glycerol-3-Phosphate Dehydrogenase, GPD1, from Sacharomyces Cerevisiae | | Descriptor: | Glycerol-3-phosphate dehydrogenase [NAD(+)] 1 | | Authors: | Aparicio, D, Munmun, N, Carpena, X, Fita, I, Loewen, P. | | Deposit date: | 2012-06-05 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase (GPD1) from Saccharomyces cerevisiae at 2.45A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4G32
 
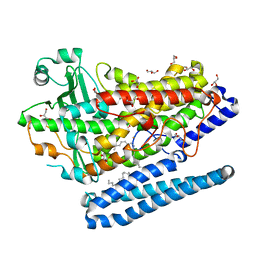 | | Crystal Structure of a Phospholipid-Lipoxygenase Complex from Pseudomonas aeruginosa at 1.75A (P21212) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, 15S-LIPOXYGENASE, FE (II) ION, ... | | Authors: | Carpena, X, Garreta, A, Val-Moraes, S.P, Garcia-Fernandez, Q, Fita, I. | | Deposit date: | 2012-07-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa.
Faseb J., 27, 2013
|
|
1P7Y
 
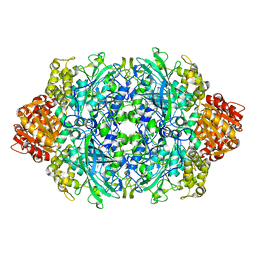 | | Crystal structure of the D181A variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1OHB
 
 | | Acetylglutamate kinase from Escherichia coli complexed with ADP and sulphate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OH9
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP, N-acetyl-L-glutamate and the transition-state mimic AlF4- | | Descriptor: | ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1P81
 
 | | Crystal structure of the D181E variant of catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1OHA
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP and N-acetyl-L-glutamate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1QWS
 
 | | Structure of the D181N variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1P7Z
 
 | | Crystal structure of the D181S variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1P80
 
 | | Crystal structure of the D181Q variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1QWL
 
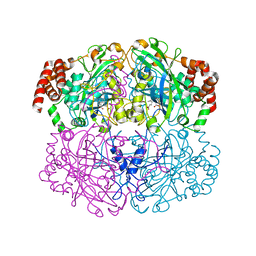 | | Structure of Helicobacter pylori catalase | | Descriptor: | AZIDE ION, KatA catalase, OXYGEN MOLECULE, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Obenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
1QWM
 
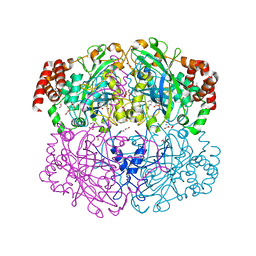 | | Structure of Helicobacter pylori catalase with formic acid bound | | Descriptor: | AZIDE ION, FORMIC ACID, KatA catalase, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Odenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
1QMY
 
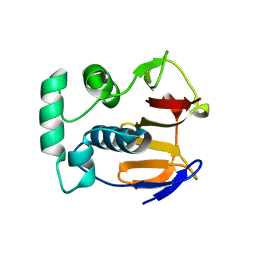 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
2WE5
 
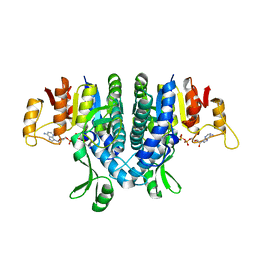 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | Authors: | Ramon-Maiques, S, Marina, A, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2WE4
 
 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | Descriptor: | CARBAMATE KINASE 1, SULFATE ION | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
6B9B
 
 | |
5L02
 
 | | S324T variant of B. pseudomallei KatG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
7BWM
 
 | |
6YRK
 
 | |
8A9B
 
 | |
8A9A
 
 | |
4HHH
 
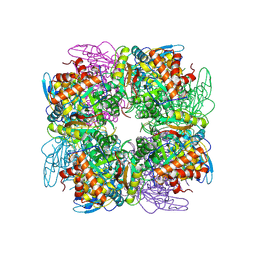 | | Structure of Pisum sativum Rubisco | | Descriptor: | RIBULOSE-1,5-DIPHOSPHATE, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Loewen, P.C, Didychuk, A.L, Switala, J, Loewen, M.C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4CAT
 
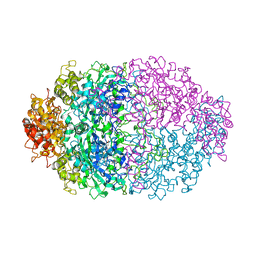 | | THREE-DIMENSIONAL STRUCTURE OF CATALASE FROM PENICILLIUM VITALE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Melik-Adamyan, W.R, Barynin, V.V, Vagin, A.A, Grebenko, A.I. | | Deposit date: | 1983-02-24 | | Release date: | 1983-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution.
J.Mol.Biol., 188, 1986
|
|
9FCH
 
 | |
