3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPL
 
 | | APO Structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPP
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
6GMU
 
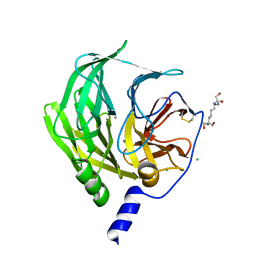 | | Serum paraoxonase-1 by directed evolution with the L69G/H134R/F222S/T332S mutations | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2018-05-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition.
Mol.Biol.Evol., 37, 2020
|
|
6FQD
 
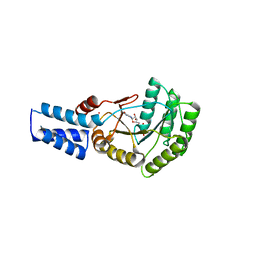 | | Escherichia Coli Signal Recognition Particle Receptor FtsY NGdN1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, POTASSIUM ION, Signal recognition particle receptor FtsY | | Authors: | Mrusek, D. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.10000563 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6H0A
 
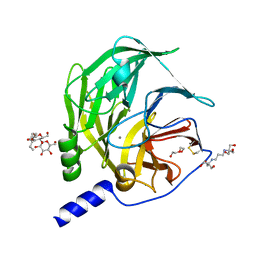 | | Serum paraoxonase-1 by directed evolution with the L69G/H115W/H134R/F222S/T332S mutations | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition.
Mol.Biol.Evol., 37, 2020
|
|
6G82
 
 | |
8R3D
 
 | | Crystal structure of Pent only at pH 8.8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta propeller | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Multivalent Calixarene Complexation of a Designed Pentameric Lectin.
Biomacromolecules, 25, 2024
|
|
8R3B
 
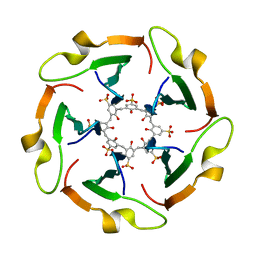 | |
8R3C
 
 | |
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
4ONV
 
 | | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate | | Descriptor: | 1,2-ETHANEDIOL, 2-KETO-3-DEOXYGLUCONATE, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate
To be Published
|
|
