4I8Q
 
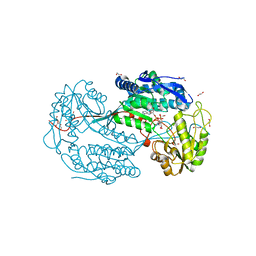 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4KPO
 
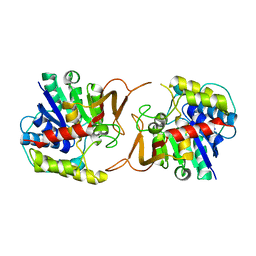 | | Plant nucleoside hydrolase - ZmNRh3 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 3 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
4K1G
 
 | |
4PXN
 
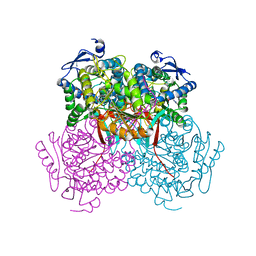 | | Structure of Zm ALDH7 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Uncharacterized protein | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
4PZ2
 
 | | Structure of Zm ALDH2-6 (RF2F) in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
4PXL
 
 | | Structure of Zm ALDH2-3 (RF2C) in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytosolic aldehyde dehydrogenase RF2C, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
4KPN
 
 | | Plant nucleoside hydrolase - PpNRh1 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 1 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
3FKB
 
 | | Structure of NDPK H122G and tenofovir-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morera, S, Chen, Y.X. | | Deposit date: | 2008-12-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
3H3K
 
 | |
3GZK
 
 | | Structure of A. Acidocaldarius Cellulase CelA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cellulase, ... | | Authors: | Morera, S, Eckert, K, Vigouroux, A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of A. acidocaldarius endoglucanase Cel9A in complex with cello-oligosaccharides: strong -1 and -2 subsites mimic cellobiohydrolase activity
J.Mol.Biol., 394, 2009
|
|
3H2W
 
 | |
3FJO
 
 | | Structure of chimeric YH CPR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Morera, S, Aigrain, L, Truan, G. | | Deposit date: | 2008-12-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase
Embo Rep., 10, 2009
|
|
1NPK
 
 | | REFINED X-RAY STRUCTURE OF DICTYOSTELIUM NUCLEOSIDE DIPHOSPHATE KINASE AT 1,8 ANGSTROMS RESOLUTION | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Lebras, G, Lascu, I, Veron, M. | | Deposit date: | 1994-07-27 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined X-ray structure of Dictyostelium discoideum nucleoside diphosphate kinase at 1.8 A resolution.
J.Mol.Biol., 243, 1994
|
|
5ITO
 
 | |
4RA1
 
 | | PBP AccA from A. tumefaciens C58 in complex with D-Glucose-2-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2014-09-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
5OVZ
 
 | | High resolution structure of the PBP NocT in complex with nopaline | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(1S)-4-carbamimidamido-1-carboxybutyl]-D-glutamic acid, ... | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
2QMH
 
 | | structure of V267F mutant HprK/P | | Descriptor: | HPr kinase/phosphorylase | | Authors: | Chaptal, V, Vincent, F, Gueguen-Chaignon, V, Poncet, S, Deutscher, J, Nessler, S, Morera, S. | | Deposit date: | 2007-07-16 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Bacterial HPr Kinase/Phosphorylase V267F Mutant Gives Insights into the Allosteric Regulation Mechanism of This Bifunctional Enzyme.
J.Biol.Chem., 282, 2007
|
|
2VED
 
 | | crystal structure of the chimerical mutant CapABK55M protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MEMBRANE PROTEIN CAPA1, ... | | Authors: | Olivares-Illana, V, Meyer, P, Gueguen-Chaignon, V, Soulat, D, Deustcher, J, Cozzone, A.J, Morera, S, Grangeasse, C, Nessler, S. | | Deposit date: | 2007-10-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Regulation Mechanism of the Tyrosine Kinase Capb from Staphylococcus Aureus.
Plos Biol., 6, 2008
|
|
3S1E
 
 | | Pro427Gln mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
6YAO
 
 | | Crystal structure of ZmCKO4a in complex with inhibitor 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
6YAP
 
 | | Crystal structure of ZmCKO4a in complex with inhibitor 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
1ANW
 
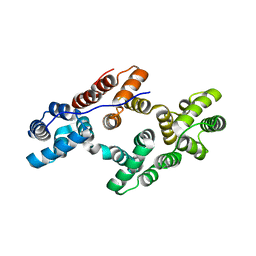 | | THE EFFECT OF METAL BINDING ON THE STRUCTURE OF ANNEXIN V AND IMPLICATIONS FOR MEMBRANE BINDING | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Lewit-Bentley, A, Morera, S, Huber, R, Bodo, G. | | Deposit date: | 1993-10-26 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The effect of metal binding on the structure of annexin V and implications for membrane binding.
Eur.J.Biochem., 210, 1992
|
|
2AK7
 
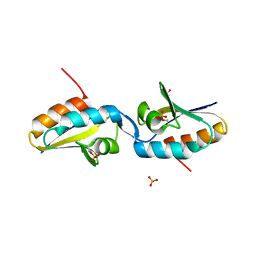 | | structure of a dimeric P-Ser-Crh | | Descriptor: | HPr-like protein crh, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Poncet, S, Lecampion, C, Lariviere, L, Meyer, P, Galinier, A, Deutscher, J, Nessler, S, Morera, S. | | Deposit date: | 2005-08-03 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a domain-swapped dimer of Ser46-phosphorylated Crh from Bacillus subtilis.
Proteins, 63, 2006
|
|
5OTC
 
 | |
