1E3C
 
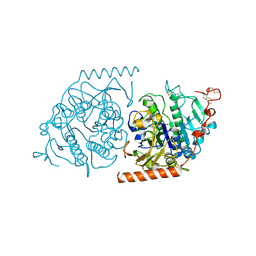 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-13 | | Release date: | 2001-03-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
2C0H
 
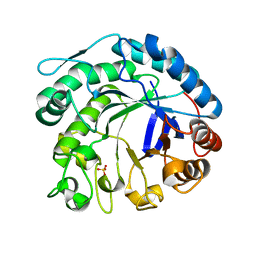 | | X-ray structure of beta-mannanase from blue mussel Mytilus edulis | | Descriptor: | MANNAN ENDO-1,4-BETA-MANNOSIDASE, SULFATE ION | | Authors: | Larsson, A.M, Anderson, L, Xu, B, Munoz, I.G, Uson, I, Janson, J.-C, Stalbrand, H, Stahlberg, J. | | Deposit date: | 2005-09-02 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-Dimensional Crystal Structure and Enzymic Characterization of Beta-Mannanase Man5A from Blue Mussel Mytilus Edulis.
J.Mol.Biol., 357, 2006
|
|
2IU1
 
 | |
7RJI
 
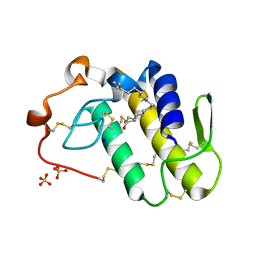 | | BthTX-II variant b, from Bothrops jararacussu venom, complexed with stearic acid | | Descriptor: | BthTX-IIb, SODIUM ION, STEARIC ACID, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | BthTX-II from Bothrops jararacussu venom has variants with different oligomeric assemblies: An example of snake venom phospholipases A 2 versatility.
Int.J.Biol.Macromol., 191, 2021
|
|
7RJZ
 
 | |
8BT9
 
 | |
6NZY
 
 | |
2DH3
 
 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ZINC ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
6FCU
 
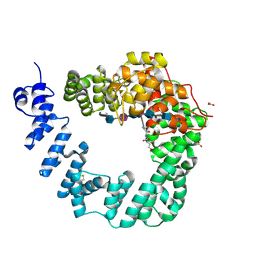 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2DH2
 
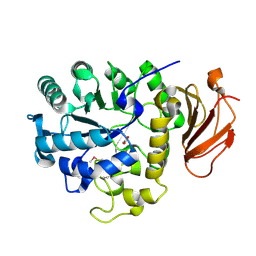 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ACETATE ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
6F62
 
 | |
8A39
 
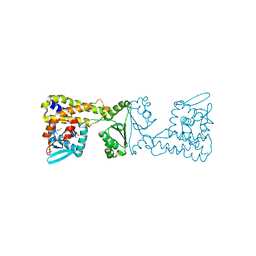 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
8AOL
 
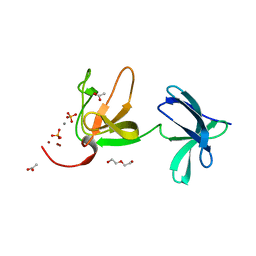 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ALU
 
 | | Crystal structure of the teichoic acid binding domain of SlpA, S-layer protein from Lactobacillus acidophilus (aa. 314-444) | | Descriptor: | PHOSPHATE ION, S-layer protein | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ALO
 
 | | Heterodimer formation of sensory domains of Vibrio cholerae regulators ToxR and ToxS | | Descriptor: | Cholera toxin transcriptional activator, Transmembrane regulatory protein ToxS | | Authors: | Gubensaek, N, Sagmeister, T, Pavkov-Keller, T, Zangger, K, Buhlheller, C, Wagner, G.E. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Vibrio cholerae's ToxRS bile sensing system.
Elife, 12, 2023
|
|
8A42
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with an unknown peptide | | Descriptor: | Oligopeptide-binding protein AmiA, Unknown peptide | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2022-06-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
4GN0
 
 | | De novo phasing of a Hamp-complex using an improved Arcimboldo method | | Descriptor: | Hamp domain of AF1503, MAGNESIUM ION | | Authors: | Hulko, M, Ursinus, A, Bar, K, Martin, J, Zeth, K, Lupas, A.N. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
5OXX
 
 | | Crystal structure of NeqN/NeqC complex from Nanoarcheaum equitans at 1.7A | | Descriptor: | NEQ068, NEQ528 | | Authors: | Aparicio, D, Perez-Luque, R, Ribo, M, Fita, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights into Subunits Assembly and the Oxyester Splicing Mechanism of Neq pol Split Intein.
Cell Chem Biol, 25, 2018
|
|
5OXW
 
 | | Structure of NeqN from Nanoarchaeum equitans | | Descriptor: | ALA-SER-GLY-SER-PHE-LYS-VAL-ILE-TYR-GLY-ASP, NEQ068 | | Authors: | Aparicio, D, Perez-Luque, R, Ribo, M, Fita, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Insights into Subunits Assembly and the Oxyester Splicing Mechanism of Neq pol Split Intein.
Cell Chem Biol, 25, 2018
|
|
5OXZ
 
 | | Crystal Structure of NeqN/NeqC complex from Nanoarcheaum equitans at 1.2A | | Descriptor: | NEQ068, NEQ528 | | Authors: | Aparicio, D, Perez-Luque, R, Ribo, M, Fita, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Insights into Subunits Assembly and the Oxyester Splicing Mechanism of Neq pol Split Intein.
Cell Chem Biol, 25, 2018
|
|
6OJB
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
8RNZ
 
 | |
6V8R
 
 | | Proteinase K Determined by MicroED Phased by ARCIMBOLDO_SHREDDER | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Richards, L.S, Martynowycz, M.W, Sawaya, M.R, Millan, C. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Fragment-based determination of a proteinase K structure from MicroED data using ARCIMBOLDO_SHREDDER
Acta Crystallogr.,Sect.D, 76, 2020
|
|
