7DZ4
 
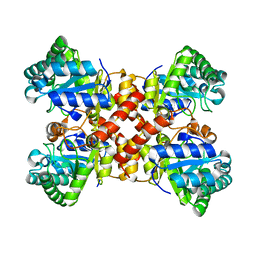 | | Crystal structures of D-allulose 3-epimerase with D-tagatose from Sinorhizobium fredii | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ6
 
 | | Crystal structures of D-allulose 3-epimerase with D-allulose from Sinorhizobium fredii | | Descriptor: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
6JO3
 
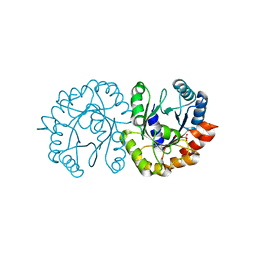 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6KZA
 
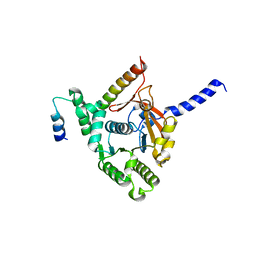 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
7CFA
 
 | |
7CO1
 
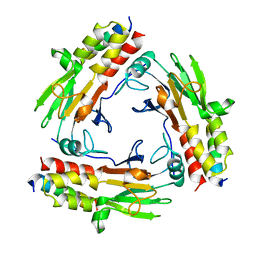 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
1V5Y
 
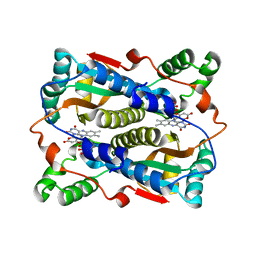 | | Binding of coumarins to NAD(P)H:FMN oxidoreductase | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, FLAVIN MONONUCLEOTIDE, Major NAD(P)H-flavin oxidoreductase | | Authors: | Kobori, T, Koike, H, Sasaki, H, Zenno, S, Saigo, K, Tanokura, M. | | Deposit date: | 2003-11-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of coumarins to NAD(P)H:FMN oxidoreductase
To be Published
|
|
1V3Z
 
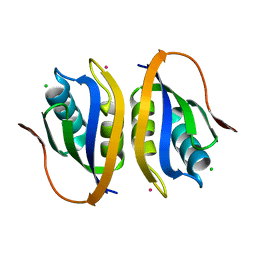 | |
1V46
 
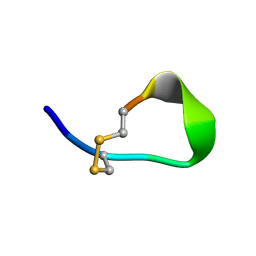 | |
1V5Z
 
 | | Binding of coumarins to NAD(P)H:FMN oxidoreductase | | Descriptor: | (2E)-3-(2-HYDROXYPHENYL)ACRYLIC ACID, FLAVIN MONONUCLEOTIDE, Major NAD(P)H-flavin oxidoreductase | | Authors: | Kobori, T, Koike, H, Sasaki, H, Zenno, S, Saigo, K, Tanokura, M. | | Deposit date: | 2003-11-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of coumarins to NAD(P)H:FMN oxidoreductase
To be Published
|
|
4YOW
 
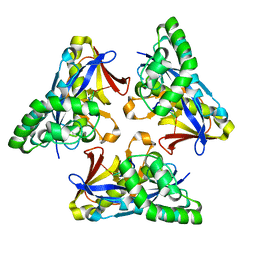 | |
4YOU
 
 | |
4YOR
 
 | |
1X25
 
 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
4YOT
 
 | |
5YPX
 
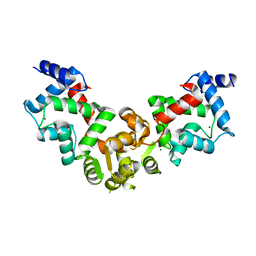 | | Crystal structure of calaxin with magnesium | | Descriptor: | Calaxin, MAGNESIUM ION | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-11-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
1VDI
 
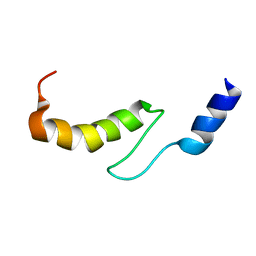 | | Solution structure of actin-binding domain of troponin in Ca2+-free state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDJ
 
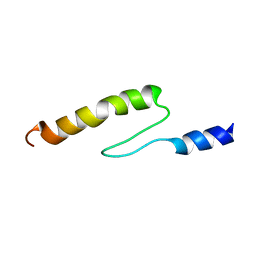 | | Solution structure of actin-binding domain of troponin in Ca2+-bound state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
4YOV
 
 | |
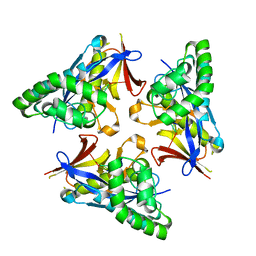
4YOY
 
 | |
4YOX
 
 | |
1Y49
 
 | |
5B0H
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKOCYTE CELL-DERIVED CHEMOTAXIN 2 | | Descriptor: | Leukocyte cell-derived chemotaxin-2, SULFATE ION, ZINC ION | | Authors: | Zheng, H, Miyakawa, T, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-10-29 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Human Leukocyte Cell-derived Chemotaxin 2 (LECT2) Reveals a Mechanistic Basis of Functional Evolution in a Mammalian Protein with an M23 Metalloendopeptidase Fold
J.Biol.Chem., 291, 2016
|
|
5X9A
 
 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
4HE7
 
 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
