4FLF
 
 | | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.15A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Sinha, M, Kaur, P, Gupta, M.N, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
4KMK
 
 | | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution
To be Published
|
|
4KL4
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Bhushan, A, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution
To be Published
|
|
4KWN
 
 | | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution
To be Published
|
|
4GLB
 
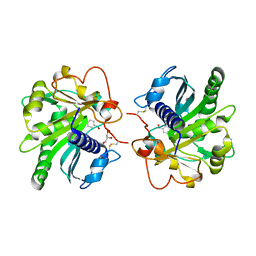 | | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution | | Descriptor: | 4-nitrobenzaldehyde, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution
TO BE PUBLISHED
|
|
4GI1
 
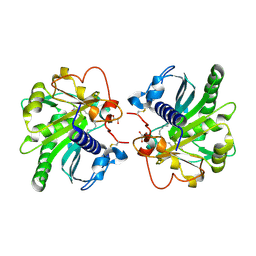 | | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution | | Descriptor: | 16-hydroxyhexadecanoic acid, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution
To be published
|
|
4Q7N
 
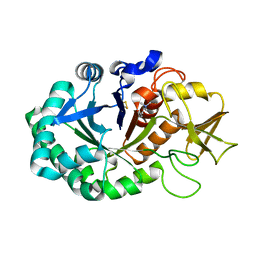 | | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, N,N,N-trimethyl-4-oxobutan-1-aminium | | Authors: | Chaudhary, A, Tyagi, T.K, Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution
To be Published
|
|
4QD3
 
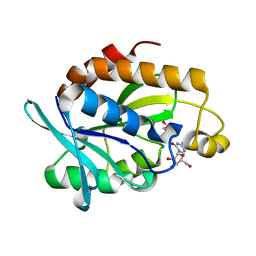 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with 5-azacytidine at 1.89 Angstrom resolution | | Descriptor: | 4-amino-1-(beta-D-ribofuranosyl)-1,3,5-triazin-2(1H)-one, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4QJQ
 
 | | Crystal structure of goat lactoperoxidase in complex with octopamine at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, ... | | Authors: | Singh, R.P, Kushwaha, G.S, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of goat lactoperoxidase in complex with octopamine at 2.1 Angstrom resolution
To be Published
|
|
4Q22
 
 | | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-04-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution
To be Published
|
|
4EMF
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with 7n-methyl-8-hydroguanosine-5-p-diphosphate at 1.77 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamini, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
4Q9F
 
 | | Crystal structure of type 1 ribosome inactivating protein from Momordica balsamina in complex with guanosine mono phosphate at 1.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Kushwaha, G.S, Pandey, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of type 1 ribosome inactivating protein from Momordica balsamina in complex with guanosine mono phosphate at 1.75 Angstrom resolution
To be Published
|
|
4QBK
 
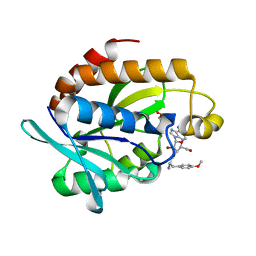 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with amino acyl-tRNA analogue at 1.77 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-[(O-methyl-L-tyrosyl)amino]adenosine, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4Q8S
 
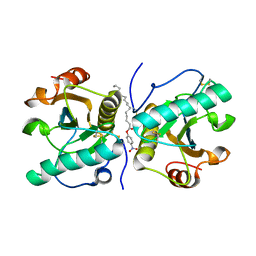 | | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrophenyl hexadecanoate, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution
To be Published
|
|
4Q9E
 
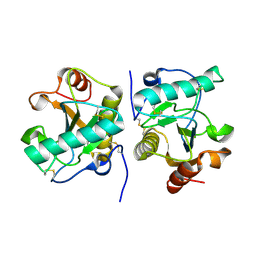 | | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution
to be published
|
|
4QAJ
 
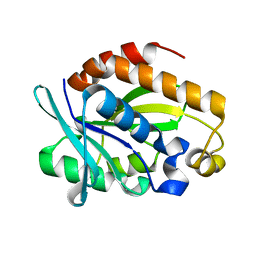 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4QMC
 
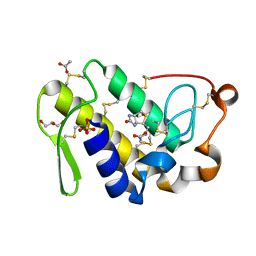 | | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution | | Descriptor: | ACETATE ION, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution
To be published
|
|
4PTM
 
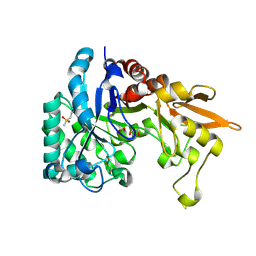 | | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution
To be Published
|
|
4QT4
 
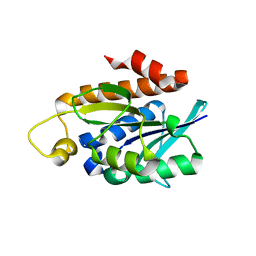 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
3OIH
 
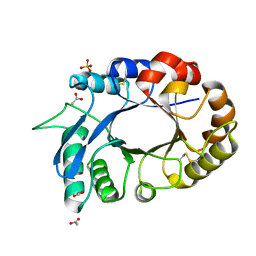 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
3N31
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Perbandt, M, Betzel, C, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution
To be Published
|
|
3QJI
 
 | | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Ribosome inactivating protein | | Authors: | Kumar, M, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-29 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution
TO BE PUBLISHED
|
|
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
4LT4
 
 | | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution
To be Published
|
|
3TDF
 
 | | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution | | Descriptor: | 2-KETOBUTYRIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaushik, S, Sinha, M, Kaur, P, Tewari, R, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution
To be Published
|
|
