5JUG
 
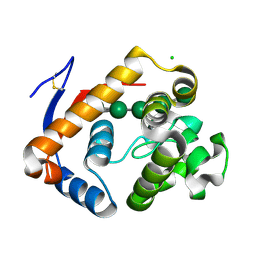 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
7P90
 
 | |
5JU9
 
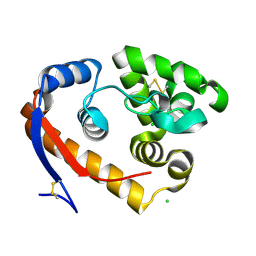 | | Structure of a beta-1,4-mannanase, SsGH134, in complex with Man3. | | Descriptor: | CHLORIDE ION, beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
7PBF
 
 | |
5KQ6
 
 | |
5KQH
 
 | |
5KQ3
 
 | |
5KQ0
 
 | |
7P25
 
 | |
6Q7J
 
 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8M
 
 | | GH10 endo-xylanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6QE8
 
 | | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-5-[(2~{S},3~{R},4~{S},5~{R})-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexane-1,2,3,4-tetrol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Wu, L, Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-01-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q7I
 
 | | GH3 exo-beta-xylosidase (XlnD) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8N
 
 | | GH10 endo-xylanase in complex with xylobiose epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{S},5~{R})-cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
8BJO
 
 | |
8BXZ
 
 | |
8C3X
 
 | |
6YWF
 
 | |
8P6O
 
 | |
8PED
 
 | |
8PC8
 
 | | Crystal structure of Paradendryphiella salina PL7C alginate lyase soaked with hexamannuronic acid | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Wilknes, C. | | Deposit date: | 2023-06-10 | | Release date: | 2023-07-12 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
8PCX
 
 | |
8PDT
 
 | |
8PC3
 
 | |
9HLI
 
 | | Influenza Neuraminidase in complex with N-Acyl Oseltamivir inhibitor | | Descriptor: | (3~{S},4~{R},5~{R},6~{R})-4-acetamido-3-azanyl-6-(butanoylamino)-5-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Moran, E, Davies, G. | | Deposit date: | 2024-12-04 | | Release date: | 2025-12-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Oseltamivir aziridines are neuraminidase imaging agents and potent anti-influenza agents
To Be Published
|
|
