7WQW
 
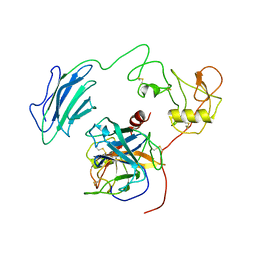 | | Structure of Active-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WR7
 
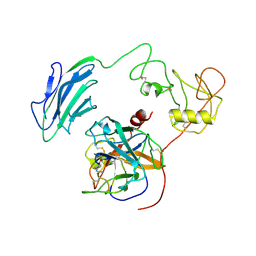 | | Structure of Inhibited-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Enteropeptidase catalytic light chain, ... | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WQX
 
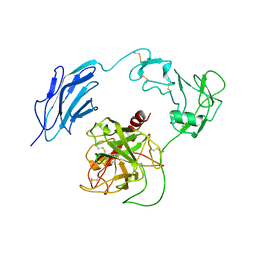 | | Structure of Inactive-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7YT9
 
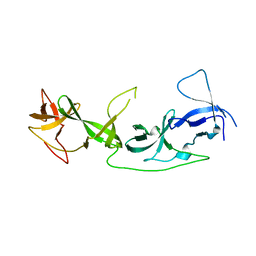 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
6H1D
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH | | Descriptor: | HemK methyltransferase family member 2, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | Deposit date: | 2018-07-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1TW4
 
 | | Crystal Structure of Chicken Liver Basic Fatty Acid Binding Protein (Bile Acid Binding Protein) Complexed With Cholic Acid | | Descriptor: | CHOLIC ACID, Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
1TVQ
 
 | | Crystal Structure of Apo Chicken Liver Basic Fatty Acid Binding Protein (or Bile Acid Binding Protein) | | Descriptor: | Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
