1PN5
 
 | | NMR structure of the NALP1 Pyrin domain (PYD) | | Descriptor: | NACHT-, LRR- and PYD-containing protein 2 | | Authors: | Hiller, S, Kohl, A, Fiorito, F, Herrmann, T, Wider, G, Tschopp, J, Grutter, M.G, Wuthrich, K. | | Deposit date: | 2003-06-12 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain
Structure, 11, 2003
|
|
1QM2
 
 | |
1RDU
 
 | | NMR STRUCTURE OF A PUTATIVE NIFB PROTEIN FROM THERMOTOGA (TM1290), WHICH BELONGS TO THE DUF35 FAMILY | | Descriptor: | conserved hypothetical protein | | Authors: | Etezady-Esfarjani, T, Herrmann, T, Peti, W, Klock, H.E, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determination of the Hypothetical Protein TM1290 from Thermotoga Maritima using Automated NOESY Analysis.
J.Biomol.NMR, 29, 2004
|
|
1SBO
 
 | | Solution Structure of putative anti sigma factor antagonist from Thermotoga maritima (TM1442) | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjaini, T, Placzek, W.J, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
2MSN
 
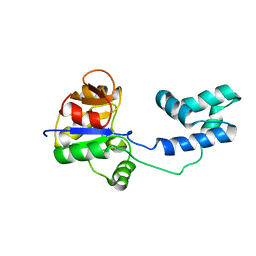 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU1
 
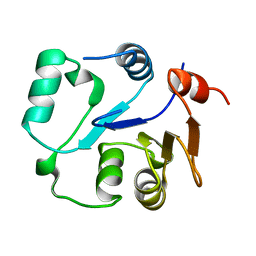 | | NMR structure of the core domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
2MCA
 
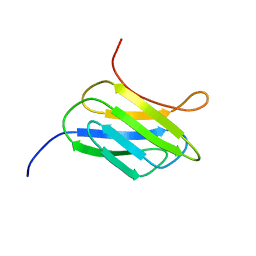 | | NMR structure of the protein YP_002937094.1 from Eubacterium rectale | | Descriptor: | Uncharacterized protein | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_002937094.1 from Eubacterium rectale
To be Published
|
|
2MW1
 
 | |
2N1M
 
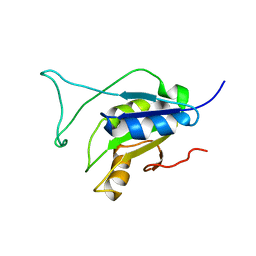 | |
2ME0
 
 | |
2M3D
 
 | |
2MXW
 
 | |
2MWM
 
 | |
2MQC
 
 | |
2MQD
 
 | |
2MCT
 
 | | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus | | Descriptor: | Uncharacterized protein | | Authors: | Martin, B.T, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus.
To be Published
|
|
2M26
 
 | |
2MVB
 
 | |
2MXV
 
 | |
2LXI
 
 | |
2MQB
 
 | |
2N2F
 
 | | Solution NMR structure of Dynorphin 1-13 bound to Kappa Opioid Receptor | | Descriptor: | Dynorphin A(1-13) | | Authors: | O'Connor, C, White, K, Doncescu, N, Didenko, T, Roth, B.L, Czaplicki, G, Stevens, R.C, Wuthrich, K, Milon, A. | | Deposit date: | 2015-05-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
