6AI0
 
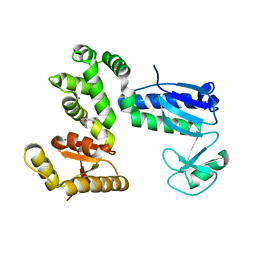 | | Structure of the 328-692 fragment of FlhA (orthorhombic form) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Flagellar biosynthesis protein FlhA | | Authors: | Ogawa, Y, Kinoshita, M, Minamino, T, Imada, K. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Substrate Specificity Switch Mechanism of the Type III Protein Export Apparatus.
Structure, 27, 2019
|
|
8GZV
 
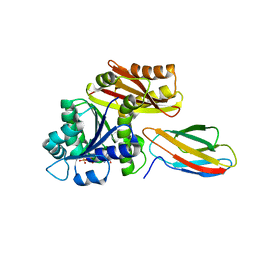 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
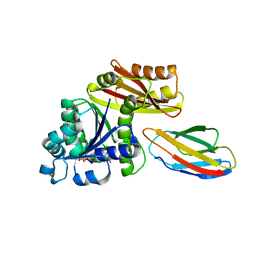 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
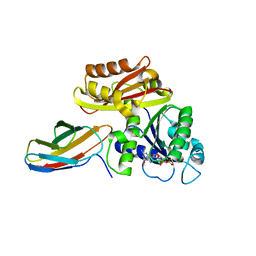 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
4A61
 
 | | ParM from plasmid R1 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | van den Ent, F, Moller-Jensen, J, Gayathri, P, Lowe, J. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
4A62
 
 | |
7CTN
 
 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | Descriptor: | Flagellar biosynthesis protein FlhA | | Authors: | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
7CIK
 
 | | Structure of the 58-213 fragment of FliF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Flagellar M-ring protein | | Authors: | Takekawa, T, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Two Distinct Conformations in 34 FliF Subunits Generate Three Different Symmetries within the Flagellar MS-Ring.
Mbio, 12, 2021
|
|
8XID
 
 | |
8YF6
 
 | |
8YF8
 
 | |
8YF9
 
 | |
8YF7
 
 | |
7WST
 
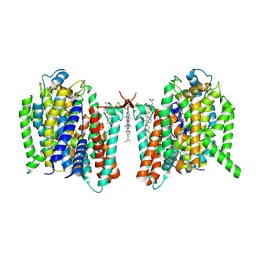 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]azetidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7WSU
 
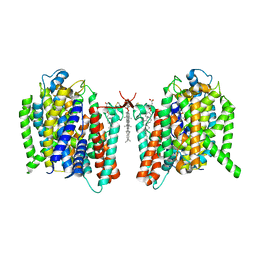 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-PDMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]pyrrolidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7WSR
 
 | | Cryo-EM structure of the barley Yellow stripe 1 transporter | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Iron-phytosiderophore transporter | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7Y7I
 
 | | chicken KNL2 in complex with the CENP-A nucleosome | | Descriptor: | Chains: I, Chains: J, Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Jiang, H, Makino, F, Fukagawa, T. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2.
Biorxiv, 2022
|
|
2D4W
 
 | |
7BW2
 
 | | Crystal Structure of Cyanobacterial PSI Monomer from T.elongatus at 6.5 A Resolution | | Descriptor: | Photosystem I 4.8K protein, Photosystem I P700 chlorophyll a apoprotein A1, Photosystem I P700 chlorophyll a apoprotein A2, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Eithar, E.M, Mian, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
1RMV
 
 | | RIBGRASS MOSAIC VIRUS, FIBER DIFFRACTION | | Descriptor: | RIBGRASS MOSAIC VIRUS COAT PROTEIN, RIBGRASS MOSAIC VIRUS RNA | | Authors: | Wang, H, Stubbs, G. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-16 | | Method: | FIBER DIFFRACTION (2.9 Å) | | Cite: | Molecular dynamics in refinement against fiber diffraction data.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
7BY0
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with the phosphorylated CENP-C | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
7BXT
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
2D4U
 
 | | Crystal Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr | | Descriptor: | Methyl-accepting chemotaxis protein I | | Authors: | Imada, K, Tajima, H, Namba, K, Sakuma, M, Homma, M, Kawagishi, I. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in the bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 2011
|
|
7UTI
 
 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM BOUND STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
