8BV7
 
 | |
5VWI
 
 | |
5VWK
 
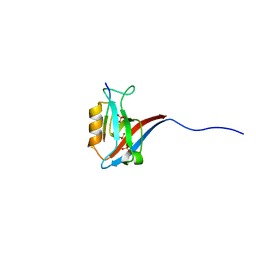 | | Crystal structure of human Scribble PDZ1:Beta-PIX complex | | Descriptor: | Beta-PIX, Protein scribble homolog, SULFATE ION | | Authors: | Lim, K.Y.B, Kvansakul, M. | | Deposit date: | 2017-05-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor beta-PIX.
J. Biol. Chem., 292, 2017
|
|
5VMN
 
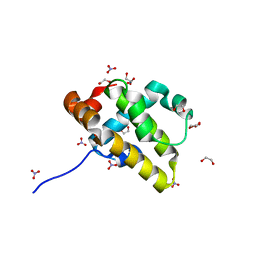 | | Crystal structure of grouper iridovirus GIV66 | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, NITRATE ION | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
4PW9
 
 | |
4PWA
 
 | |
4PW3
 
 | |
