4B3G
 
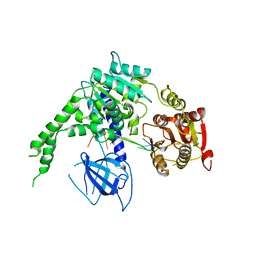 | | crystal structure of Ighmbp2 helicase in complex with RNA | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, RNA (5'-(AP*AP*AP*AP*AP*AP*AP*AP*AP)-3') | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
5OK1
 
 | |
5OK2
 
 | |
5NFE
 
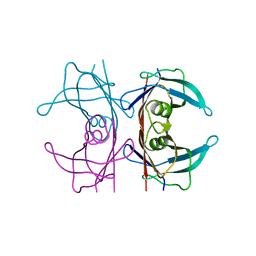 | | Neutron structure of human transthyretin (TTR) T119M mutant at room temperature to 1.85A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Ostermann, A, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.853 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
5OJZ
 
 | |
5NFW
 
 | | Neutron structure of human transthyretin (TTR) S52P mutant at room temperature to 1.8A resolution (quasi-Laue) | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
5OK0
 
 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
8Q4S
 
 | |
8QOB
 
 | |
4N0A
 
 | | Crystal structure of Lsm2-3-Pat1C complex from Saccharomyces cerevisiae | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Wu, D.H. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Cell Res., 24, 2014
|
|
4NG2
 
 | |
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
4Z4Y
 
 | | Crystal structure of GII.10 P domain in complex with 7.5mM B antigen (trisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4S
 
 | | Crystal structure of GII.10 P domain in complex with 150mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4V
 
 | | Crystal structure of GII.10 P domain in complex with 19mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4Z
 
 | | Crystal structure of GII.10 P domain in complex with 30mM B antigen (trisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4U
 
 | | Crystal structure of GII.10 P domain in complex with 37.5mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4B3F
 
 | | crystal structure of Ighmbp2 helicase | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, PHOSPHATE ION | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
4EMK
 
 | | Crystal structure of SpLsm5/6/7 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5, U6 snRNA-associated Sm-like protein LSm6, U6 snRNA-associated Sm-like protein LSm7 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4EMH
 
 | | Crystal structure of SpLsm4 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm4 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4EMG
 
 | | Crystal structure of SpLsm3 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
