7WZN
 
 | | PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
8YEE
 
 | | Cryo-EM structure of in vitro reconstituted Light-harvesting complex II trimer | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Seki, S, Miyata, T, Tanaka, H, Namba, K, Kurisu, G, Fujii, R. | | Deposit date: | 2024-02-22 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structure-based validation of recombinant light-harvesting complex II.
Pnas Nexus, 3, 2024
|
|
8Y15
 
 | | Cryo-EM structure of Light-harvesting complex from Spinacia oleracea | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Seki, S, Miyata, T, Tanaka, H, Namba, K, Kurisu, G, Fujii, R. | | Deposit date: | 2024-01-23 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structure-based validation of recombinant light-harvesting complex II.
Pnas Nexus, 3, 2024
|
|
9KKH
 
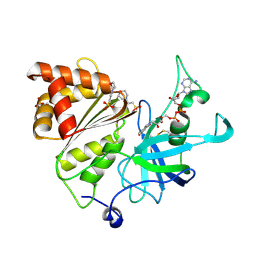 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Reduced form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KK7
 
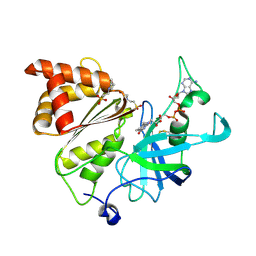 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Reduced form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKC
 
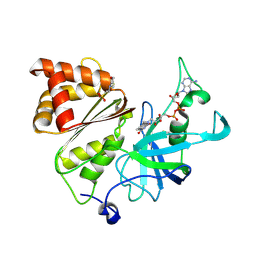 | | Neutron structure of Ferredoxin-NADP+ reductase from maize root -Oxidized form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9KKG
 
 | | High resolution structure of Ferredoxin-NADP+ reductase from maize root - Oxidized form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-11-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9K9M
 
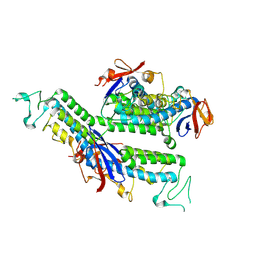 | | X-ray structure of FBB18 from Chlamydomonas reinhardtii. | | Descriptor: | Flagellar associated protein | | Authors: | Sahashi, Y, Tanaka, H, Shimo-Kon, R, Yamamoto, R, Kurisu, G, Kon, T. | | Deposit date: | 2024-10-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chlamydomonas FBB18 is a ubiquitin-like protein essential for the cytoplasmic preassembly of various ciliary dyneins.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
7W2J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7WLM
 
 | | The Cryo-EM structure of siphonaxanthin chlorophyll a/b type light-harvesting complex II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Seki, S, Nakaniwa, T, Castro-Hartmann, P, Sader, K, Kawamoto, A, Tanaka, H, Qian, P, Kurisu, G, Fujii, R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into blue-green light utilization by marine green algal light harvesting complex II at 2.78 angstrom.
Bba Adv, 2, 2022
|
|
7WSQ
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
5BX5
 
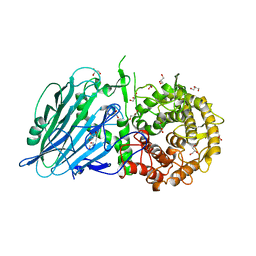 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Hua, Y, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BX4
 
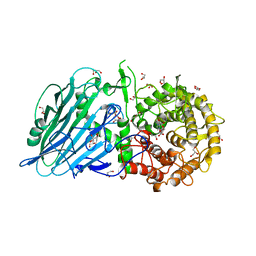 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with Glucoimidazole | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUCOIMIDAZOLE, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BX3
 
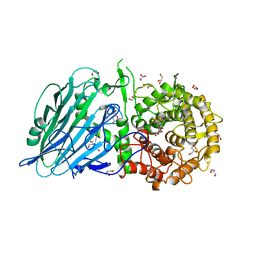 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BVU
 
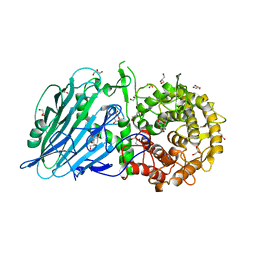 | | Crystal structure of Thermoanaerobacterium xylolyticum GH116 beta-glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-05 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2).
Acs Chem.Biol., 11, 2016
|
|
5BX2
 
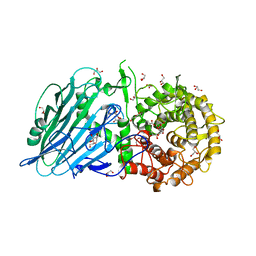 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
6J13
 
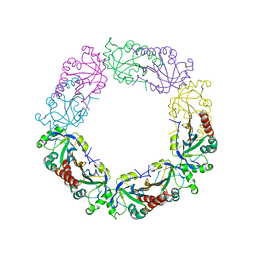 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
6L4P
 
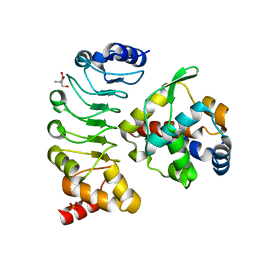 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
7Y5Y
 
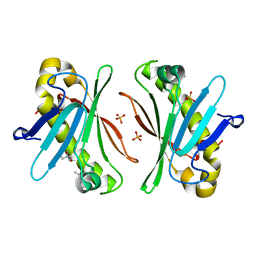 | | X-ray Structure of Stay-Green (SGR) from Anaerolineae bacterium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Dey, D, Nishijima, M, Kurisu, G, Tanaka, H, Ito, H. | | Deposit date: | 2022-06-18 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and reaction mechanism of a bacterial Mg-dechelatase homolog from the Chloroflexi Anaerolineae.
Protein Sci., 31, 2022
|
|
2EC7
 
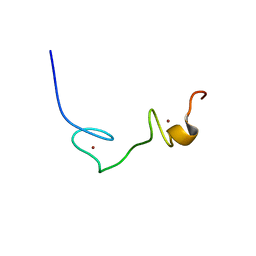 | | Solution Structure of Human Immunodificiency Virus Type-2 Nucleocapsid Protein | | Descriptor: | Gag polyprotein (Pr55Gag), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Tanaka, T, Endoh, H, Tanaka, H, Miyauchi, E, Komatsu, H, Kohno, T, Maeda, T. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7
Biochemistry, 48, 2009
|
|
2E1X
 
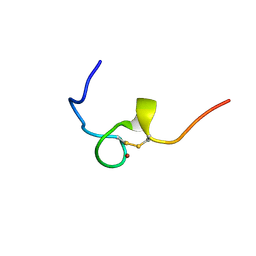 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Miyauchi, E, Tanaka, H, Endoh, H, Komatsu, H, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-11-03 | | Release date: | 2007-06-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural role of the secondary active domain of HIV-2 NCp8 in multi-functionality
Biochem.Biophys.Res.Commun., 358, 2007
|
|
7XHX
 
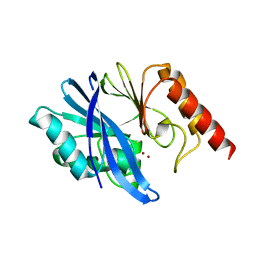 | | Crystal structure of metallo-beta-lactamase IMP-6 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XHW
 
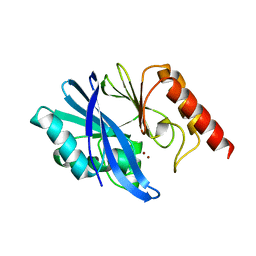 | | Crystal structure of metallo-beta-lactamase IMP-1 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
