6KCC
 
 | |
6K1Y
 
 | |
6LZO
 
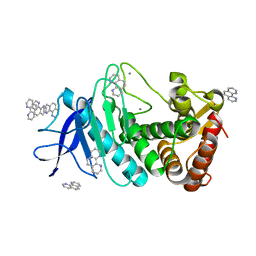 | | Thermolysin with 1,10-phenanthroline | | Descriptor: | 1,10-PHENANTHROLINE, CALCIUM ION, Thermolysin | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
6LZN
 
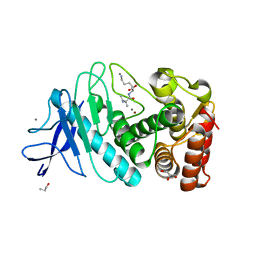 | | Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
6LL2
 
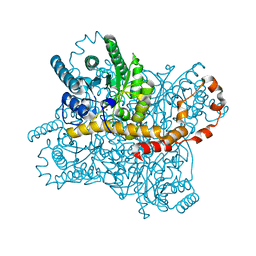 | |
6LL3
 
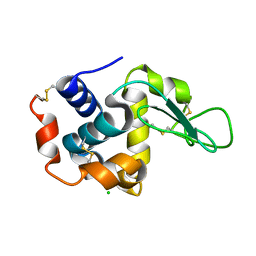 | |
3H17
 
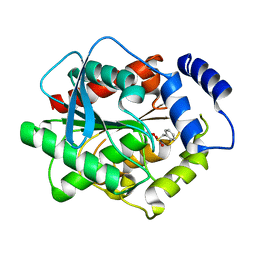 | | Crystal structure of EstE5-PMSF (I) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3GVY
 
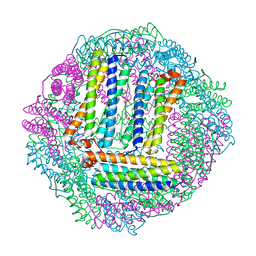 | |
3II1
 
 | |
3K6K
 
 | |
9LPU
 
 | | Crystal structure of tKeima at pH 4.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2025-01-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima.
Molecules, 30, 2025
|
|
3H18
 
 | | Crystal structure of EstE5-PMSF (II) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
8IH0
 
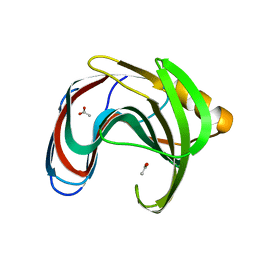 | | Crystal structure of GH11 from Thermoanaerobacterium saccharolyticum | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase | | Authors: | Nam, K.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structural analysis of the endo-1,4-beta-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification.
Sci Rep, 13, 2023
|
|
8IH1
 
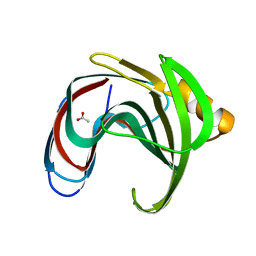 | |
3G6N
 
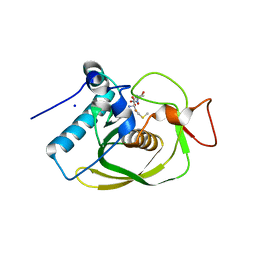 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
7CVM
 
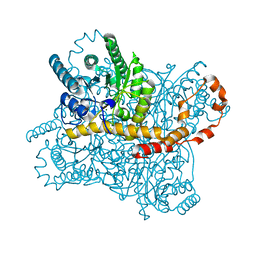 | |
7CVJ
 
 | |
7CVK
 
 | |
7CVL
 
 | |
3DNM
 
 | |
3FAK
 
 | |
3CMJ
 
 | |
3CMD
 
 | | Crystal structure of peptide deformylase from VRE-E.faecium | | Descriptor: | FE (III) ION, MALONATE ION, Peptide deformylase, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2008-03-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure.
Proteins, 74, 2009
|
|
3FW6
 
 | |
8WDG
 
 | |
