1V0Y
 
 | |
2YG8
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA-3-methyladenine glycosidase II, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
2VRN
 
 | | The structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily | | Descriptor: | MAGNESIUM ION, PROTEASE I | | Authors: | Fioravanti, E, Dura, M.A, Lascoux, D, Micossi, E, McSweeney, S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily.
Biochemistry, 47, 2008
|
|
2YG9
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, DNA-3-methyladenine glycosidase II, putative, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1UTH
 
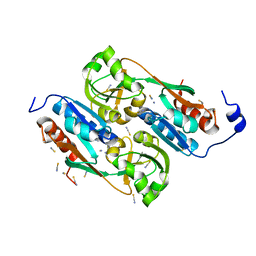 | | DntR from Burkholderia sp. strain DNT in complex with Thiocyanate | | Descriptor: | LYSR-TYPE REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Smirnova, I.A, Dian, C, Leonard, G.A, McSweeney, S, Birse, D, Brzezinski, P. | | Deposit date: | 2003-12-09 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a Bacterial Biosensor for Nitrotoluenes: The Crystal Structure of the Transcriptional Regulator Dntr
J.Mol.Biol., 340, 2004
|
|
1UTB
 
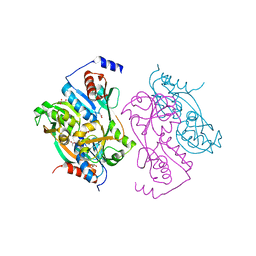 | | DntR from Burkholderia sp. strain DNT | | Descriptor: | ACETATE ION, GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Smirnova, I.A, Dian, C, Leonard, G.A, McSweeney, S, Birse, D, Brzezinski, P. | | Deposit date: | 2003-12-05 | | Release date: | 2004-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of a Bacterial Biosensor for Nitrotoluenes: The Crystal Structure of the Transcriptional Regulator Dntr
J.Mol.Biol., 340, 2004
|
|
1USP
 
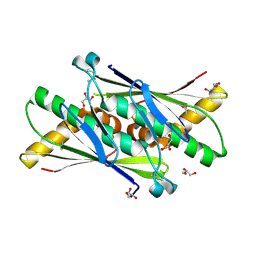 | | Organic Hydroperoxide Resistance Protein from Deinococcus radiodurans | | Descriptor: | GLYCEROL, ORGANIC HYDROPEROXIDE RESISTANCE PROTEIN | | Authors: | Meunier-Jamin, C, Kapp, U, Leonard, G, McSweeney, S. | | Deposit date: | 2003-11-27 | | Release date: | 2004-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Organic Hydroperoxide Resistance Protein from Deinococcus Radiodurans: Do Conformational Changes Facilitate Recycling of the Redox Disulfide?
J.Biol.Chem., 279, 2004
|
|
1SHQ
 
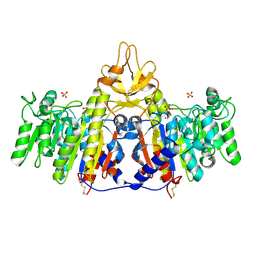 | | Crystal structure of shrimp alkaline phosphatase with magnesium in M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SHN
 
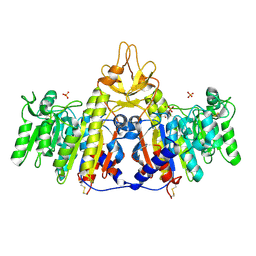 | | Crystal structure of shrimp alkaline phosphatase with phosphate bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZGT
 
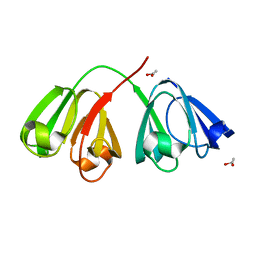 | | Structure of hydrogenated rat gamma E crystallin in H2O | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIR
 
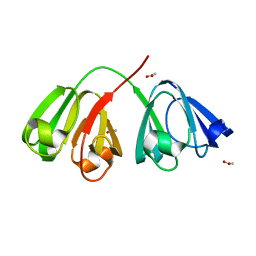 | | Deuterated gammaE crystallin in H2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BHY
 
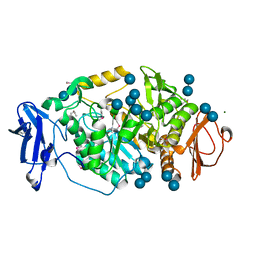 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with trehalose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2C2F
 
 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-28 | | Release date: | 2006-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
2BHU
 
 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MALTOOLIGOSYLTREHALOSE TREHALOHYDROLASE, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BHZ
 
 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with maltose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BOO
 
 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2U
 
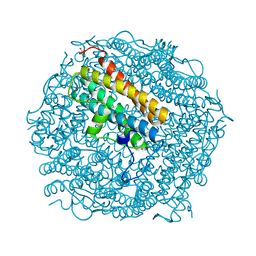 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, SULFATE ION, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-30 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
1ZIQ
 
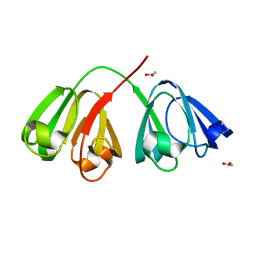 | | Deuterated gammaE crystallin in D2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2P
 
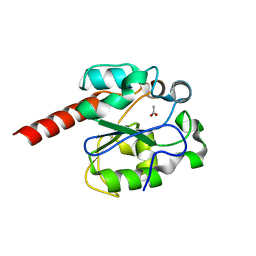 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
1ZIE
 
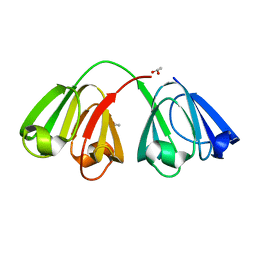 | | Hydrogenated gammaE crystallin in D2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2Q
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans. Inactive mutant Asp93Ala. | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
1AH5
 
 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6Y8G
 
 | | selenomethionine derivative of ferulic acid esterase (FAE) | | Descriptor: | CADMIUM ION, Endo-1,4-beta-xylanase Y, GLYCEROL | | Authors: | von Stetten, D, Mueller-Dieckmann, C, Carpentier, P, Flot, D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
4YTA
 
 | | BOND LENGTH ANALYSIS OF ASP, GLU AND HIS RESIDUES IN TRYPSIN AT 1.2A RESOLUTION | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3UNX
 
 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
