3JSC
 
 | | CcdBVfi-FormI-pH7.0 | | Descriptor: | CcdB, SULFATE ION | | Authors: | De Jonge, N, Buts, L, Loris, R. | | Deposit date: | 2009-09-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
4GC1
 
 | |
4GC2
 
 | |
7AEX
 
 | |
8CJ5
 
 | |
8CJ8
 
 | |
8CO2
 
 | |
4ELY
 
 | | CCDBVFI:GYRA14EC | | Descriptor: | CHLORIDE ION, CcdB, DNA gyrase subunit A, ... | | Authors: | De Jonge, N, Simic, R, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
4HOH
 
 | | RIBONUCLEASE T1 (THR93ALA MUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
5J9I
 
 | |
5JAA
 
 | |
5JA9
 
 | | Crystal structure of the HigB2 toxin in complex with Nb6 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 6, SULFATE ION, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
5JA8
 
 | | Crystal structure of the HigB2 toxin in complex with Nb2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
5MI8
 
 | | Structure of the phosphomimetic mutant of EF-Tu T383E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
5MI9
 
 | | Structure of the phosphomimetic mutant of the elongation factor EF-Tu T62E | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
5MI3
 
 | | Structure of phosphorylated translation elongation factor EF-Tu from E. coli | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
5MJE
 
 | | Crystal structure of the HigB2 toxin in complex with Nb8 | | Descriptor: | Cytotoxic translational repressor of toxin-antitoxin stability system, DI(HYDROXYETHYL)ETHER, Nanobody 8, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
4ELZ
 
 | | CCDBVFI:GYRA14VFI | | Descriptor: | CcdB, DNA gyrase subunit A, GLYCEROL | | Authors: | De Jonge, N, Simic, M, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
4Z33
 
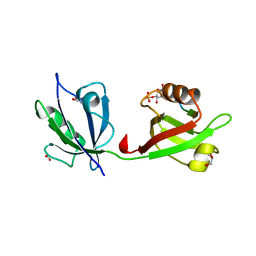 | | Crystal structure of the syntenin PDZ1 and PDZ2 tandem in complex with the Frizzled 7 C-terminal fragment and PIP2 | | Descriptor: | ACETATE ION, D-MYO-INOSITOL-4,5-BISPHOSPHATE, GLYCEROL, ... | | Authors: | Egea-Jimenez, A.L, Gallardo, R, Garcia-Pino, A, Ivarsson, Y, Wawrzyniak, A.M, Kashyap, R, Loris, R, Schymkowitz, J, Rousseau, F, Zimmermann, P. | | Deposit date: | 2015-03-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the syntenin PDZ1 and PDZ2 tandem in complex with the Frizzled 7 C-terminal fragment and PIP2
To Be Published
|
|
2ENR
 
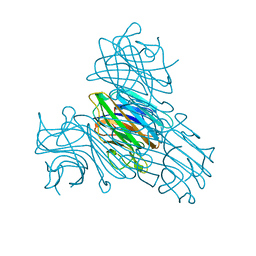 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH CADMIUM HAVING A CADMIUM ION BOUND IN BOTH THE S1 SITE AND THE S2 SITE | | Descriptor: | CADMIUM ION, CONCANAVALIN A | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1LUL
 
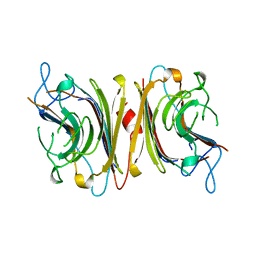 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1G7Y
 
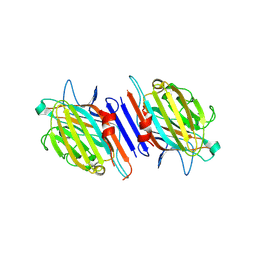 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
1CES
 
 | | CRYSTALS OF DEMETALLIZED CONCANAVALIN A SOAKED WITH ZINC HAVE A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-02-15 | | Release date: | 1997-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1BJQ
 
 | | THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH ADENINE | | Descriptor: | ADENINE, CALCIUM ION, LECTIN, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1RXE
 
 | | ArsC complexed with MNB | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Arsenate reductase, PERCHLORATE ION, ... | | Authors: | Messens, J, Van Molle, I, Vanhaesebrouck, P, Limbourg, M, Van Belle, K, Wahni, K, Martins, J.C, Loris, R, Wyns, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-06-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a triple mutant of pI258 arsenate reductase from Staphylococcus aureus and its 5-thio-2-nitrobenzoic acid adduct.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
