6KEN
 
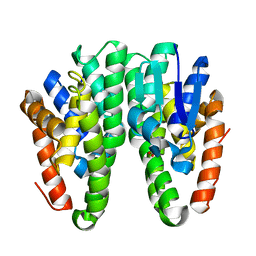 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
5XWM
 
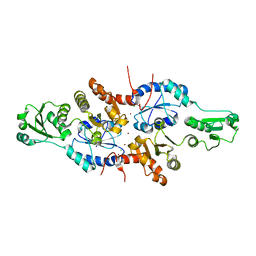 | | human ERp44 zinc-bound form | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44, ZINC ION | | Authors: | Watanabe, S, Harayama, M, Inaba, K. | | Deposit date: | 2017-06-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Zinc regulates ERp44-dependent protein quality control in the early secretory pathway.
Nat Commun, 10, 2019
|
|
6IGG
 
 | | Crystal structure of FT condition 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGJ
 
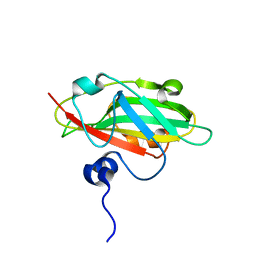 | | Crystal structure of FT condition 4 | | Descriptor: | MAGNESIUM ION, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6JJU
 
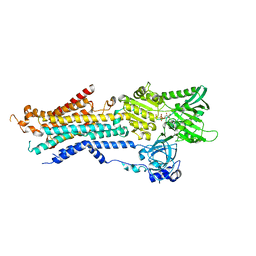 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
6IGI
 
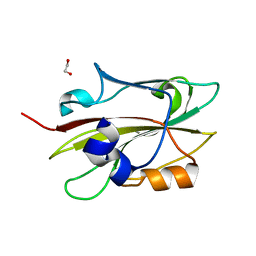 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGH
 
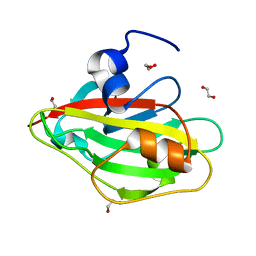 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
2KP1
 
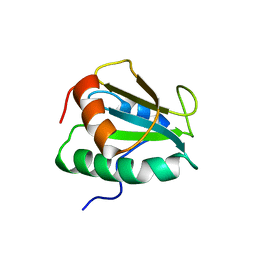 | |
2KP2
 
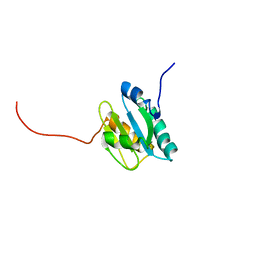 | |
5AUQ
 
 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
