4LWH
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
7YTY
 
 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
7YTW
 
 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
4R3M
 
 | | Crystal structure of Human Hsp90 with JR9 | | Descriptor: | Heat shock protein HSP 90-alpha, N~3~-benzyl-2-[(6-bromo-1,3-benzodioxol-5-yl)methyl]imidazo[1,2-a]pyrazine-3,8-diamine | | Authors: | Li, J, Yang, M, Ren, J, Xiong, B, He, J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multi-substituted 8-aminoimidazo[1,2-a]pyrazines by Groebke-Blackburn-Bienayme reaction and their Hsp90 inhibitory activity.
Org.Biomol.Chem., 13, 2015
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H8C
 
 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8B
 
 | | Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form | | Descriptor: | Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8A
 
 | | Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
3JBM
 
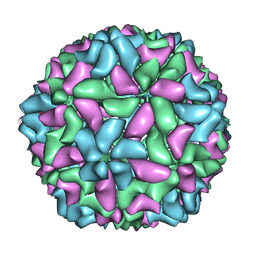 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
4L8Z
 
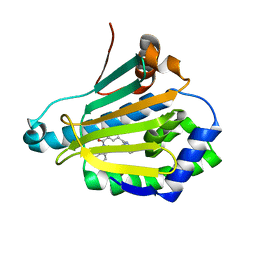 | | Crystal structure of Human Hsp90 with RL1 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](3,4-dihydroisoquinolin-2(1H)-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
9JVP
 
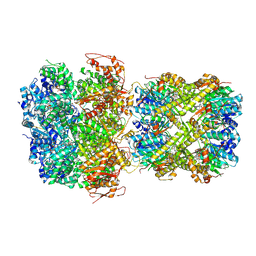 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
9JVZ
 
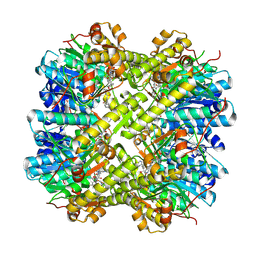 | | CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, W, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
9IV9
 
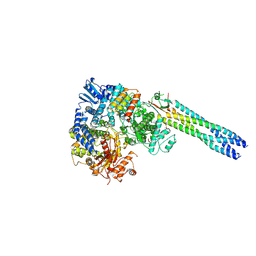 | | Cryo-EM structure of a truncated Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
9IVA
 
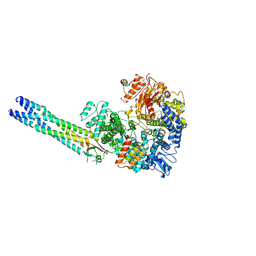 | | Cryo-EM structure of the full-length Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
4L93
 
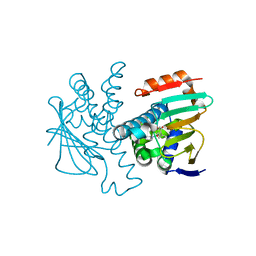 | | Crystal structure of Human Hsp90 with S36 | | Descriptor: | 3,4-dihydroisoquinolin-2(1H)-yl[2,4-dihydroxy-5-(propan-2-yl)phenyl]methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Crystal structure of Human Hsp90 with S36
To be Published
|
|
4L90
 
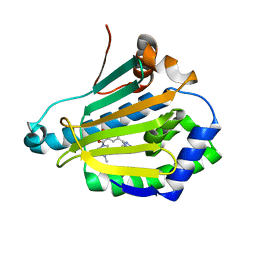 | | Crystal structure of Human Hsp90 with RL3 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Human Hsp90 with RL3
to be published
|
|
5CDI
 
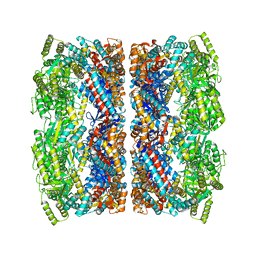 | | Chloroplast chaperonin 60b1 of Chlamydomonas | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Zhou, H, Yu, F, Gao, F, He, J, Liu, C. | | Deposit date: | 2015-07-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural insight into the cooperation of chloroplast chaperonin subunits
Bmc Biol., 14, 2016
|
|
2I99
 
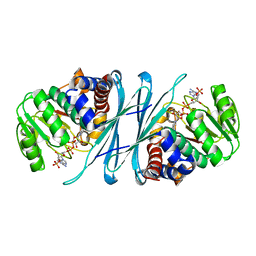 | | Crystal structure of human Mu_crystallin at 2.6 Angstrom | | Descriptor: | Mu-crystallin homolog, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cheng, Z, Sun, L, He, J, Gong, W. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human {micro}-crystallin complexed with NADPH
Protein Sci., 16, 2007
|
|
7YD2
 
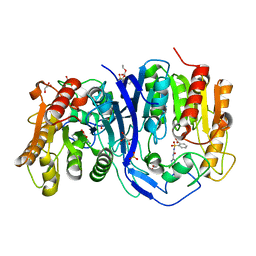 | | SulE_P44R_S209A | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7Y0L
 
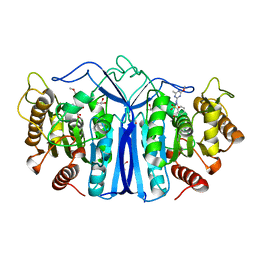 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7XM0
 
 | | Crystal structure of Sau3AI-C and DNA substrate complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*CP*AP*TP*G)-3'), MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Liu, Y, Yu, F, He, J. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Sau3AI with and without bound DNA suggest a self-activation-based DNA cleavage mechanism.
Structure, 31, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
