1RVK
 
 | | Crystal structure of enolase AGR_L_2751 from Agrobacterium Tumefaciens | | Descriptor: | MAGNESIUM ION, isomerase/lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Zencheck, W, Millikin, C, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-14 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activites in the Enolase superfamily: 1.7 A crystal structure of the hypothetical protein MR.GI-17937161 from Agrobacterium tumefaciens
To be Published
|
|
4FX8
 
 | | Crystal structure of the mutant Q185A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9411 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4GC3
 
 | | Crystal structure of L-HISTIDINOL PHOSPHATE PHOSPHATASE (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and sulfate | | Descriptor: | L-HISTIDINOL PHOSPHATE PHOSPHATASE, SULFATE ION, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
4IP4
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg | | Descriptor: | CARBON DIOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with mg
To be Published
|
|
4IP5
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate | | Descriptor: | (2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate
To be Published
|
|
4GK8
 
 | | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and L-histidinol arsenate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidinol-phosphatase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
4HCH
 
 | | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-tartrate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Isomerase/lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-tartrate
To be Published
|
|
4GYF
 
 | | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN, L-histidinol and phosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidinol-phosphatase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN, L-histidinol and phosphate
To be Published
|
|
4ICM
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis | | Descriptor: | 5-carboxyvanillate decarboxylase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis
To be Published
|
|
4HCD
 
 | | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium | | Descriptor: | CHLORIDE ION, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Millikin, C, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium
To be Published
|
|
4J8L
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT protein YhfS from Escherichia coli | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, NICKEL (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfs from escherichia coli
To be Published
|
|
4HCL
 
 | | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate
To be Published
|
|
4K1W
 
 | | Crystal structure of the A314P mutant of mannonate dehydratase from novosphingobium aromaticivorans complexed with mg and d-mannonate | | Descriptor: | 1,3-PROPANDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-04-06 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Crystal structure of the A314P mutant of mannonate
dehydratase from novosphingobium aromaticivorans complexed with mg
and d-mannonate
To be Published
|
|
4KBX
 
 | | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx from escherichia coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-04-24 | | Release date: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx from escherichia coli
To be Published
|
|
3HK5
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK8
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3IJI
 
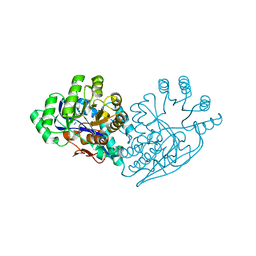 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ID7
 
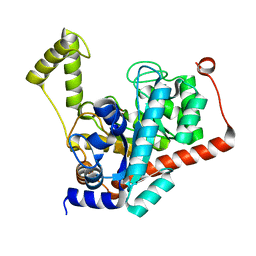 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3IJQ
 
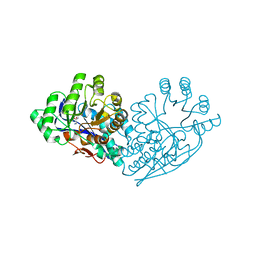 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IJL
 
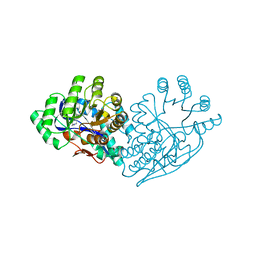 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Pro-D-Glu; nonproductive substrate binding. | | Descriptor: | D-GLUTAMIC ACID, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JW7
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ile-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, ISOLEUCINE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K1G
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ser-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JVA
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Imker, H, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JTE
 
 | | Crystal structure of response regulator receiver domain Protein from clostridium thermocellum | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of response regulator receiver domain
Protein from clostridium thermocellum
To be Published
|
|
3K5X
 
 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
