5KS1
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, ... | | Authors: | Ussin, N, Abdulsalam, R.W, Offermann, L.R, Chruszcz, M. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
5KRV
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus in complex Arginine | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ARGININE, ... | | Authors: | Ussin, N, Abdulsalam, R.W, Magee, P, Chruszcz, M. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
7S2H
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
7S2P
 
 | | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, SULFATE ION | | Authors: | Carey, S.M, Nettles, R.B, Daneshian, L, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN
To Be Published
|
|
4DGT
 
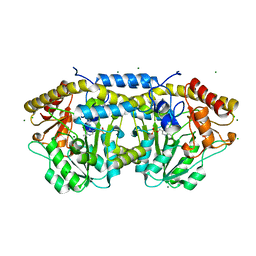 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 crystallized with magnesium formate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
7S2N
 
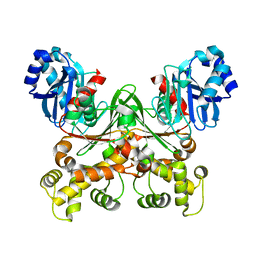 | |
4DQ6
 
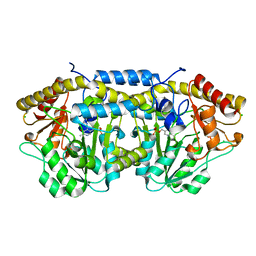 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Putative pyridoxal phosphate-dependent transferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
4EUY
 
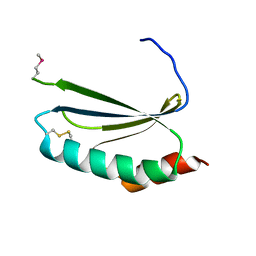 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
4X9U
 
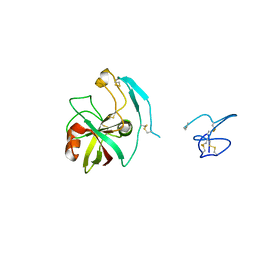 | | Crystal structure of the kiwifruit allergen Act d 5 | | Descriptor: | Kiwellin | | Authors: | Offermann, L.R, Perdue, M.L, Giangrieco, I, Tamburrini, M, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin.
J.Agric.Food Chem., 63, 2015
|
|
4KLV
 
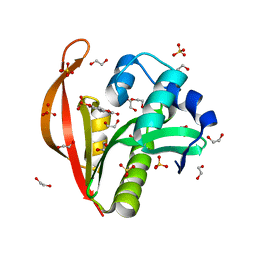 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 4-methylumbelliferyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, 4-methylumbelliferyl phosphate, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOT
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOX
 
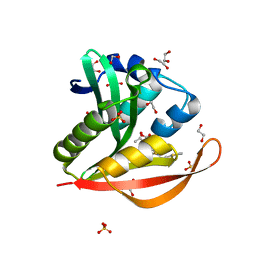 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefalotin | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOTHIN, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOR
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 7-aminocephalosporanic acid | | Descriptor: | 1,2-ETHANEDIOL, 7-aminocephalosporanic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOV
 
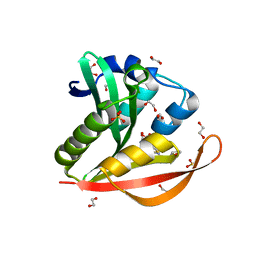 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefuroxime | | Descriptor: | (6R,7R)-3-[(carbamoyloxy)methyl]-7-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOS
 
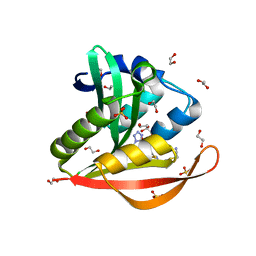 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefmetazole | | Descriptor: | (6R,7S)-7-({[(cyanomethyl)sulfanyl]acetyl}amino)-7-methoxy-3-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-8-oxo-5-thia -1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4X9K
 
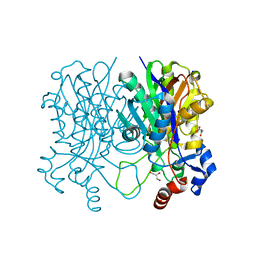 | | Beta-ketoacyl-acyl carrier protein synthase III-2 (FabH2)(C113A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Chordia, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4KLW
 
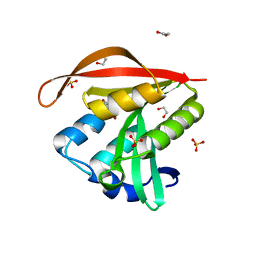 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 2-(aminocarbonyl)benzoate | | Descriptor: | 1,2-ETHANEDIOL, 2-carbamoylbenzoic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOU
 
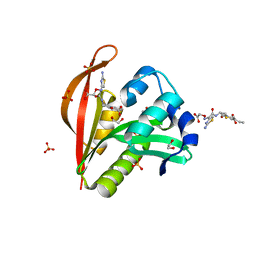 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefixime | | Descriptor: | (6R,7R)-7-({(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-[(carboxymethoxy)imino]acetyl}amino)-3-ethenyl-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4XUW
 
 | | Structure of the hazelnut allergen, Cor a 8 | | Descriptor: | Non-specific lipid-transfer protein, PHOSPHATE ION | | Authors: | Offermann, L.R, Perdue, M.L, Bublin, M, Pfeifer, S, Dubiela, P, Hoffmann-Sommergruber, K, Chruszcz, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.
J.Agric.Food Chem., 63, 2015
|
|
6V8L
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | Ara h 8 allergen isoform, SULFATE ION, icosanoic acid | | Authors: | Pote, S, Offermann, L.R, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
6V8M
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Ara h 8 allergen isoform, SULFATE ION | | Authors: | Pote, S, Offermann, L.R, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
6V8H
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | Ara h 8 allergen isoform, SULFATE ION | | Authors: | Offermann, L.R, Pote, S, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
6V8J
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | Ara h 8 allergen isoform, SODIUM ION, SULFATE ION | | Authors: | Offermann, L.R, Pote, S, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
6V8S
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Ara h 8 allergen isoform, SULFATE ION | | Authors: | Pote, S, Offermann, L.R, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
4XKY
 
 | | Structure of dihydrodipicolinate synthase from the commensal bacterium Bacteroides thetaiotaomicron at 2.1 A resolution | | Descriptor: | ACETATE ION, Dihydrodipicolinate synthase, SODIUM ION | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrodipicolinate synthase from the commensal bacterium Bacteroides thetaiotaomicron at 2.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
