5XMV
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS362 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-chlorophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5- carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5YG4
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS849 | | Descriptor: | 2-[1-[(3~{S})-6'-azanyl-5'-cyano-7-fluoranyl-2,2,3'-trimethyl-spiro[1~{H}-indene-3,4'-2~{H}-pyrano[2,3-c]pyrazole]-5-yl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
5YG2
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS705 | | Descriptor: | 3-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
5YG3
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS834 | | Descriptor: | (3~{S})-6'-azanyl-7-fluoranyl-2,2,3'-trimethyl-5-pyridin-4-yl-spiro[1~{H}-indene-3,4'-2~{H}-pyrano[2,3-c]pyrazole]-5'-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
5YFZ
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS626 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
7OMO
 
 | |
7OMR
 
 | |
7OMD
 
 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | Descriptor: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | Authors: | Schenkmayerova, A, Janin, Y.L, Marek, M. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
6N1G
 
 | |
7QXR
 
 | | Azacoelenterazine-bound Renilla-type luciferase (AncFT) | | Descriptor: | 3-(4-hydroxyphenyl)-8-[(4-hydroxyphenyl)methyl]-5-(phenylmethyl)-1$l^{4},4,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Fragment transplantation onto a hyperstable ancestor of haloalkane dehalogenases and Renilla luciferase (Anc-FT) | | Authors: | Marek, M, Schenkmayerova, A, Janin, Y.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
7QXQ
 
 | | Coelenteramide-bound Renilla-type luciferase (AncFT) | | Descriptor: | Fragment transplantation onto hyperstable ancestor of haloalkane dehalogenases and Renilla luciferase (Anc-FT), N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE | | Authors: | Marek, M, Schenkmayerova, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
5WYR
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
7EL2
 
 | |
7EL3
 
 | |
7DCH
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with acarbose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glycosidase, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9C
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-13 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9B
 
 | | Crystal structure of alpha-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DCG
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EHI
 
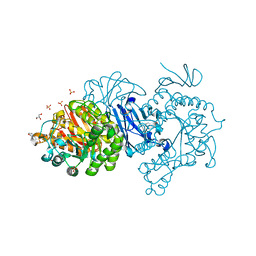 | | Crystal structure of covalent maltosyl-alpha-glucosidase intermediate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EHH
 
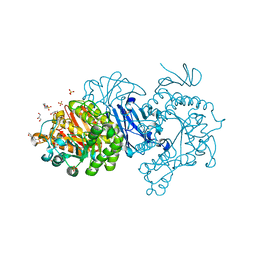 | | Crystal structure of alpha-glucosidase from Weissella cibaria BKK1 in complex with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6G7O
 
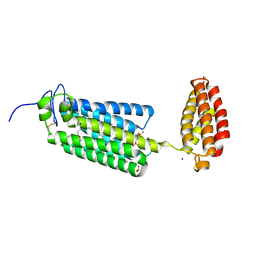 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
5KWW
 
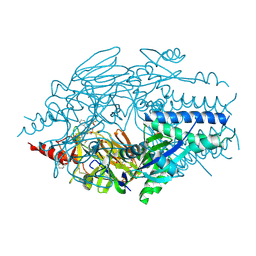 | | Crystal Structure of Inhibitor JNJ-53718678 In Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[5-chloranyl-1-(3-methylsulfonylpropyl)indol-2-yl]methyl]-1-[2,2,2-tris(fluoranyl)ethyl]imidazo[4,5-c]pyridin-2-one, ... | | Authors: | McLellan, J.S, Battles, M.B, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2016-07-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor.
Nat Commun, 8, 2017
|
|
4TN4
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with glycine and a novel pyrazolopyran 33G: (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Witschel, M.C. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT): Cocrystal Structures of Pyrazolopyrans with Potent Blood- and Liver-Stage Activities.
J.Med.Chem., 58, 2015
|
|
4TMR
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with glycine and a novel pyrazolopyran 99S: methyl 5-{3-[(4S)-6-amino-5-cyano-3-methyl-4-(propan-2-yl)-2,4-dihydropyrano[2,3-c]pyrazol-4-yl]-5-cyanophenyl}thiophene-2-carboxylate . | | Descriptor: | CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Witschel, M.C. | | Deposit date: | 2014-06-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT): Cocrystal Structures of Pyrazolopyrans with Potent Blood- and Liver-Stage Activities.
J.Med.Chem., 58, 2015
|
|
8YSB
 
 | | Crystal structure of DynA1, a putative monoxygenase from Mivromonospora chersina. | | Descriptor: | Predicted ester cyclase | | Authors: | Yan, X.F, Huang, H.W, Gao, Y.G, Liang, Z.X. | | Deposit date: | 2024-03-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Enzymatic Oxidation Cascade Converts delta-Thiolactone Anthracene to Anthraquinone in the Biosynthesis of Anthraquinone-Fused Enediynes.
Jacs Au, 4, 2024
|
|
