6Y8S
 
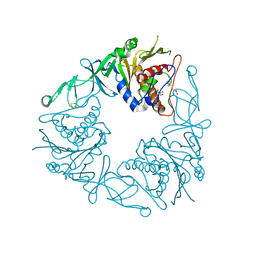 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
5BZ3
 
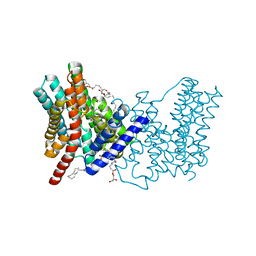 | | CRYSTAL STRUCTURE OF SODIUM PROTON ANTIPORTER NAPA IN OUTWARD-FACING CONFORMATION. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, Na(+)/H(+) antiporter | | Authors: | Coincon, M, Uzdavinys, P, Emmanuel, N, Cameron, A, Drew, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5BZ2
 
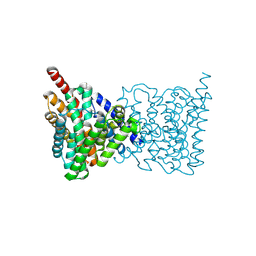 | |
1GQT
 
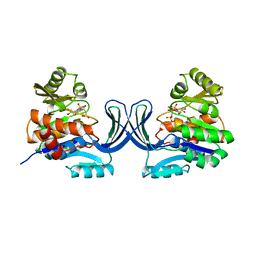 | |
2WIP
 
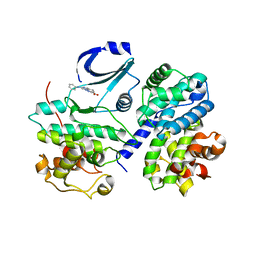 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
3AIE
 
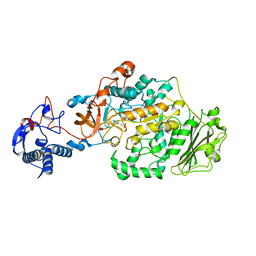 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIB
 
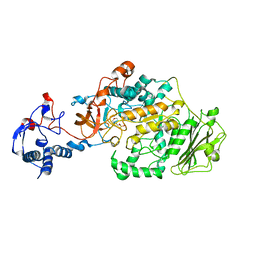 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIC
 
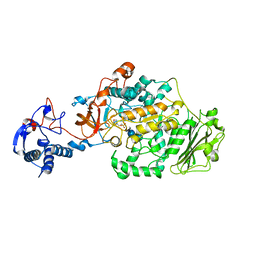 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
1BYL
 
 | |
1MNH
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNJ
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNI
 
 | |
1MNK
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
