8EQN
 
 | |
7SQJ
 
 | |
7SN4
 
 | |
7SN7
 
 | |
7SN9
 
 | |
8EJJ
 
 | | Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJE
 
 | | Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein GP1, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJF
 
 | | Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJH
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab | | Descriptor: | 12.1F Fab heavy chain, 12.1F Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
8DZK
 
 | | Dbr1 in complex with 5-mer cleavage product | | Descriptor: | FE (II) ION, RNA (5'-R(P*(G46)P*UP*GP*UP*U)-3'), RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B. | | Deposit date: | 2022-08-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product.
Biochemistry, 61, 2022
|
|
8ESC
 
 | | Structure of the Yeast NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Patel, A.B, Zukin, S.A, Nogales, E. | | Deposit date: | 2022-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and flexibility of the yeast NuA4 histone acetyltransferase complex.
Elife, 11, 2022
|
|
8F1A
 
 | | Apo KIF20A[1-565] class-1 in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Crozet, V, Ranaivoson, F.M, Houdusse, A, Sosa, H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
8F18
 
 | | Apo KIF20A[1-565] class-2 in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Crozet, V, Ranaivoson, F.M, Houdusse, A, Sosa, H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
8F5I
 
 | | SARS-CoV-2 S2 helix epitope scaffold bound by antibody DH1057.1 | | Descriptor: | DH1057.1 HC, DH1057.1 LC, Specialized acyl carrier protein, ... | | Authors: | Kapingidza, A.B, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
8F5H
 
 | |
1MCL
 
 | | PRINCIPLES AND PITFALLS in DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), N-ACETYL-D-HIS-L-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCR
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCC
 
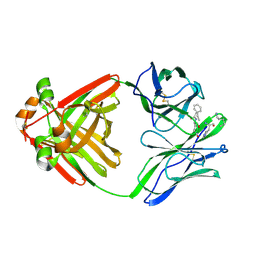 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCO
 
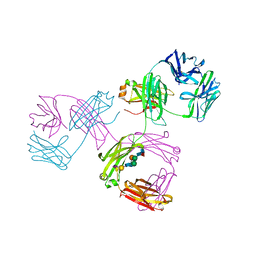 | | THREE-DIMENSIONAL STRUCTURE OF A HUMAN IMMUNOGLOBULIN WITH A HINGE DELETION | | Descriptor: | IGG1 MCG INTACT ANTIBODY (HEAVY CHAIN), IGG1 MCG INTACT ANTIBODY (LIGHT CHAIN), N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-L-gulopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guddat, L.W, Edmundson, A.B. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional structure of a human immunoglobulin with a hinge deletion.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
1MCE
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCN
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-HIS-L-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCF
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
